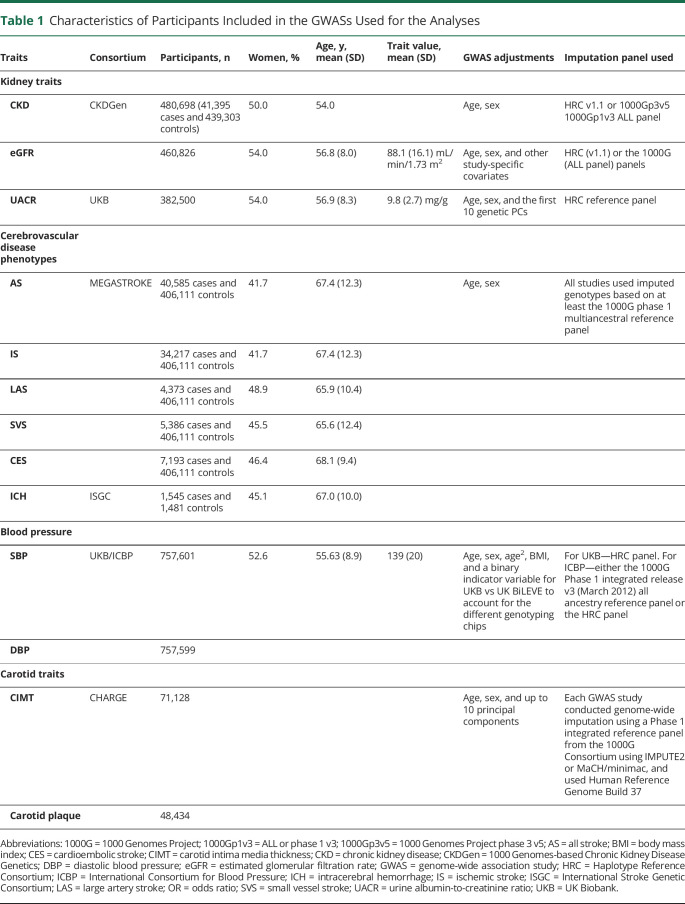
Table 1.
Characteristics of Participants Included in the GWASs Used for the Analyses
| Traits | Consortium | Participants, n | Women, % | Age, y, mean (SD) | Trait value, mean (SD) | GWAS adjustments | Imputation panel used |
| Kidney traits | |||||||
| CKD | CKDGen | 480,698 (41,395 cases and 439,303 controls) | 50.0 | 54.0 | Age, sex | HRC v1.1 or 1000Gp3v5 1000Gp1v3 ALL panel | |
| eGFR | 460,826 | 54.0 | 56.8 (8.0) | 88.1 (16.1) mL/min/1.73 m2 | Age, sex, and other study-specific covariates | HRC (v1.1) or the 1000G (ALL panel) panels | |
| UACR | UKB | 382,500 | 54.0 | 56.9 (8.3) | 9.8 (2.7) mg/g | Age, sex, and the first 10 genetic PCs | HRC reference panel |
| Cerebrovascular disease phenotypes | |||||||
| AS | MEGASTROKE | 40,585 cases and 406,111 controls | 41.7 | 67.4 (12.3) | Age, sex | All studies used imputed genotypes based on at least the 1000G phase 1 multiancestral reference panel | |
| IS | 34,217 cases and 406,111 controls | 41.7 | 67.4 (12.3) | ||||
| LAS | 4,373 cases and 406,111 controls | 48.9 | 65.9 (10.4) | ||||
| SVS | 5,386 cases and 406,111 controls | 45.5 | 65.6 (12.4) | ||||
| CES | 7,193 cases and 406,111 controls | 46.4 | 68.1 (9.4) | ||||
| ICH | ISGC | 1,545 cases and 1,481 controls | 45.1 | 67.0 (10.0) | |||
| Blood pressure | |||||||
| SBP | UKB/ICBP | 757,601 | 52.6 | 55.63 (8.9) | 139 (20) | Age, sex, age2, BMI, and a binary indicator variable for UKB vs UK BiLEVE to account for the different genotyping chips | For UKB—HRC panel. For ICBP—either the 1000G Phase 1 integrated release v3 (March 2012) all ancestry reference panel or the HRC panel |
| DBP | 757,599 | ||||||
| Carotid traits | |||||||
| CIMT | CHARGE | 71,128 | Age, sex, and up to 10 principal components | Each GWAS study conducted genome-wide imputation using a Phase 1 integrated reference panel from the 1000G Consortium using IMPUTE2 or MaCH/minimac, and used Human Reference Genome Build 37 | |||
| Carotid plaque | 48,434 |
Abbreviations: 1000G = 1000 Genomes Project; 1000Gp1v3 = ALL or phase 1 v3; 1000Gp3v5 = 1000 Genomes Project phase 3 v5; AS = all stroke; BMI = body mass index; CES = cardioembolic stroke; CIMT = carotid intima media thickness; CKD = chronic kidney disease; CKDGen = 1000 Genomes-based Chronic Kidney Disease Genetics; DBP = diastolic blood pressure; eGFR = estimated glomerular filtration rate; GWAS = genome-wide association study; HRC = Haplotype Reference Consortium; ICBP = International Consortium for Blood Pressure; ICH = intracerebral hemorrhage; IS = ischemic stroke; ISGC = International Stroke Genetic Consortium; LAS = large artery stroke; OR = odds ratio; SVS = small vessel stroke; UACR = urine albumin-to-creatinine ratio; UKB = UK Biobank.