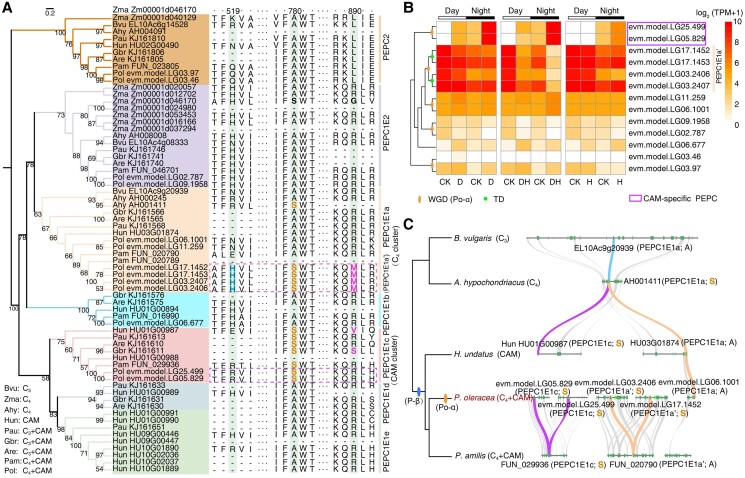
Figure 3.
Analysis of C4- and CAM-specific PEPC genes in the common purslane genome. A) Phylogenetic tree of PEPC proteins from P. oleracea (Pol, C4 + CAM), P. amilis (Pam, C4 + CAM), H. undatus (Hun, CAM), A. hypochondriacus (Ahy, C4), B. vulgaris (Bvu, C3), Z. mays (Zma, C4), and several identified PEPC proteins from Pereskia aureiflora (Pau, C3 + CAM), Anacampseros retusa (Are, C3 + CAM), and Grahamia bracteata (Gbr, C3 + CAM) in Caryophyllales. Only bootstrap values greater than 50% were shown. Branches were colored according to the PEPC subclass. A multiple sequence alignment of partially conserved amino acids was shown to the right. Sites at positions of 519 (known as position of D509 in the Kalanchoë genome), 780, and 890 (according to the numbering in Zm00001d046170) were highlighted in light-green background. Potentially functional C4- and CAM-specific PEPC proteins were highlighted in magenta and purple boxes, respectively. B) Gene expression of PEPC genes identified in P. oleracea under heat and drought treatments during the day and night. Orange ovals in the gene trees to the left represent WGD Po-α events; green dots represent TD events. CAM-specific PEPC genes were highlighted in magenta boxes. C) Synteny analysis of C4- and CAM-specific PEPC genes in the P. oleracea, P. amilis, H. undatus, A. hypochondriacus, and B. vulgaris genomes in Caryophyllales. CK, control group; D, drought group; H, heat group; DH, drought combined heat group.