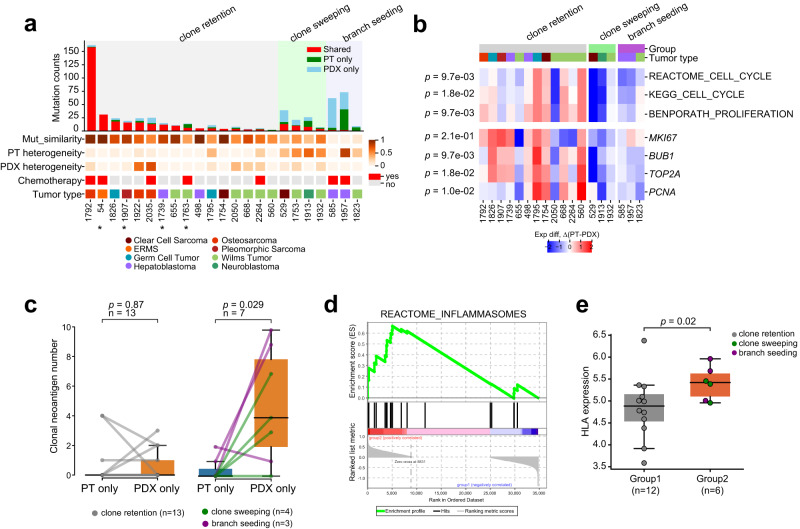
Fig. 4. Evolutionary pattern is associated with mutational similarity and antitumor immunity in PT.
a The evolutionary patterns are correlated with genetic heterogeneity of the PT. Patients are ordered by the number of shared mutations between PT and PDX within each pattern. The top bar plot shows mutation overlap for each PT/PDX pair. The bottom panel shows mutational similarity, PT genetic heterogeneity, PDX genetic heterogeneity, chemotherapy, and cancer type. Samples labeled with asterisk do not have the matched germline. b Difference in expression of proliferation markers and cell cycle signatures. Red, higher in PT; Blue, higher in PDX. The signatures were scored with ssGSEA. The p-values on the left were calculated between group 1 and group 2 using two-sided Wilcoxon rank sum test. c Changes in clonal neoantigen count between PTs and PDXs. The left panel shows the changes for group 1 samples, and the right panel shows those for group 2 samples. Each dot represents one sample. P-values were calculated with two-sided paired t test. The box represents the interquartile range (IQR), and middle line represents median. Whiskers represent values within 1.5×IQR of the upper and lower quantiles respectively. d Pathway analysis identifies inflammasome pathway higher in group 2 PTs compared with group 1 PTs (p = 0.03). P-value is reported as is from Gene Set Enrichment Analysis. e HLA genes, which encode MHC complexes on APC surfaces, are highly expressed in group 2 PTs (p = 0.02, one-sided Wilcoxon rank sum test). HLA gene expression is summarized using ssGSEA. The boxplots are interpreted similarly as in panel c. Source data are provided as a Source data file.