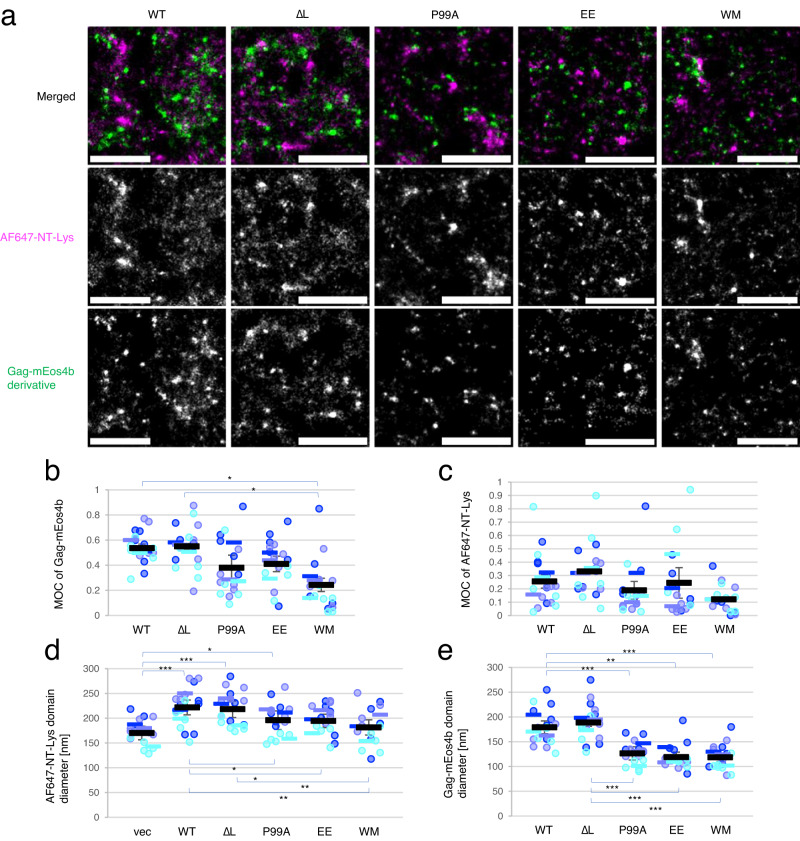
Fig. 7. Colocalization between Gag-mEos4b curvature mutants and AF647-NT-Lys and domain analysis in PALM/dSTORM.
a Representative images of Gag/Gag-mEos4b derivatives-expressing HeLa cells labeled with AF647-NT-Lys. From top to bottom: merged, AF647-NT-Lys, and Gag-mEos4b derivatives images. Bar, 1 µm. Vec, cells expressing an empty vector; WT, Gag WT/Gag WT-mEos4b; ∆L, Gag-∆L/Gag-∆L-mEos4b; P99A, Gag-P99A/Gag-P99A-mEos4b; EE, Gag-EE/Gag-EE-mEos4b; WM, Gag-WM/Gag-WM-mEos4b. b MOC values in colocalization analysis of Gag-mEos4b derivatives to AF647-NT-Lys. c MOC values of AF647-NT-Lys to Gag-mEos4b derivatives. d, e the mean diameters [nm] of AF647-NT-Lys (d) and Gag-mEos4b derivatives (e) domains in each cell, the means in each of three independent experiments, and the mean ± SEM were shown as dots and bars in three different blue colors, and black bars with the error bars (n = 18, 17, 16, 16, and 16 for WT, ∆L, P99A, EE, and WM over three experiments). *, **, and *** indicate significant differences at the significance levels of 0.05, 0.01, and 0.005 in the one-way ANOVA post-hoc Tukey test, respectively. For comparison, the data of vec, WT, and WM samples in Figs. 2c–f, 3a, c were shown again. Source data and detailed statistics for (b–e) were provided as a Source Data file.