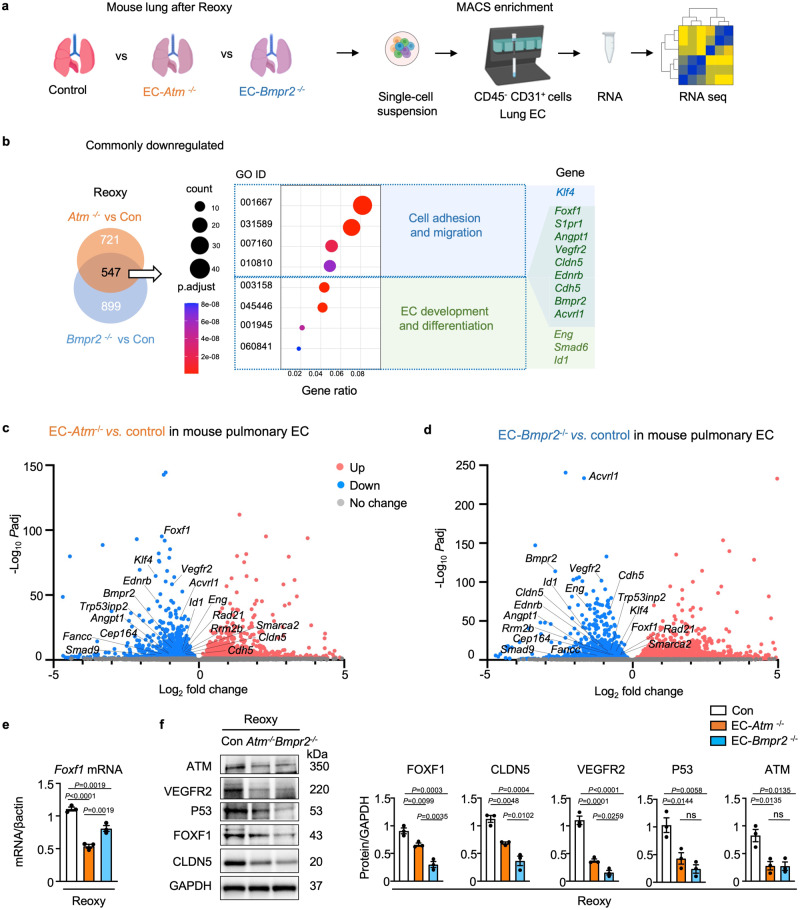
Fig. 4. Transcriptome analysis reveals that angiogenesis pathways are downregulated in Bmpr2- and Atm-deleted murine PAEC.
a Scheme of study design. The lungs of tamoxifen-inducible EC-Atm-/-, EC-Bmpr2-/- and control mice (n = 5) after reoxygenation were harvested, enzymatically digested to obtain single-cell suspensions, and pulmonary endothelial cells (CD45− CD31+ cells) were collected using magnetic beads. Total RNA of pulmonary EC from n = 5 mice was used for bulk RNA sequencing. Schema created with BioRender.com. b Venn diagram showing the number of genes downregulated following reoxygenation in lung EC of EC-Atm-/- or EC-Bmpr2-/- mice, compared to control mice [adjusted P-value (Padj) <0.05]. Gene Ontology (GO) analysis was performed for 547 commonly downregulated genes to reveal the top GO pathways. P-values were adjusted using the Benjamini–Hochberg method for multiple comparisons by one-sided Fisher’s exact test. c, d Volcano plots displaying DEGs (differentially expressed genes) following reoxygenation in EC-Atm-/- or EC-Bmpr2-/- compared to control mice (c, d respectively). Angiogenesis genes (Foxf1, Vegfr2, Acvrl1, S1pr1, Eng, Bmpr2, Id1, Klf4, Cdh5, Cldn5, Ednrb), Trp53inp2, a gene downstream of p53, and Foxf1 targeting as well as DNA repair genes (Fancc, Smarca2, Rad21, Rrm2b, Cep164) are indicated on the plots. 2-sided Wald test was performed. P-values were adjusted to get Padj using the Benjamini-Hochberg method. e Foxf1 expression in lung EC of EC-Atm-/-, EC-Bmpr2-/- and control mice following reoxygenation. n = 3 biological replicates. f Immunoblot and densitometry of FOXF1, CLDN5, VEGFR2, P53 and ATM in pulmonary EC of EC-Atm-/-, EC-Bmpr2-/- and control mice after reoxygenation. n = 3 biological replicates. e, f Bars represent mean ± S.E.M. P values determined by one-way ANOVA with Holm-Sidak posthoc test. ns, not significant. Source data are provided as a Source Data file.