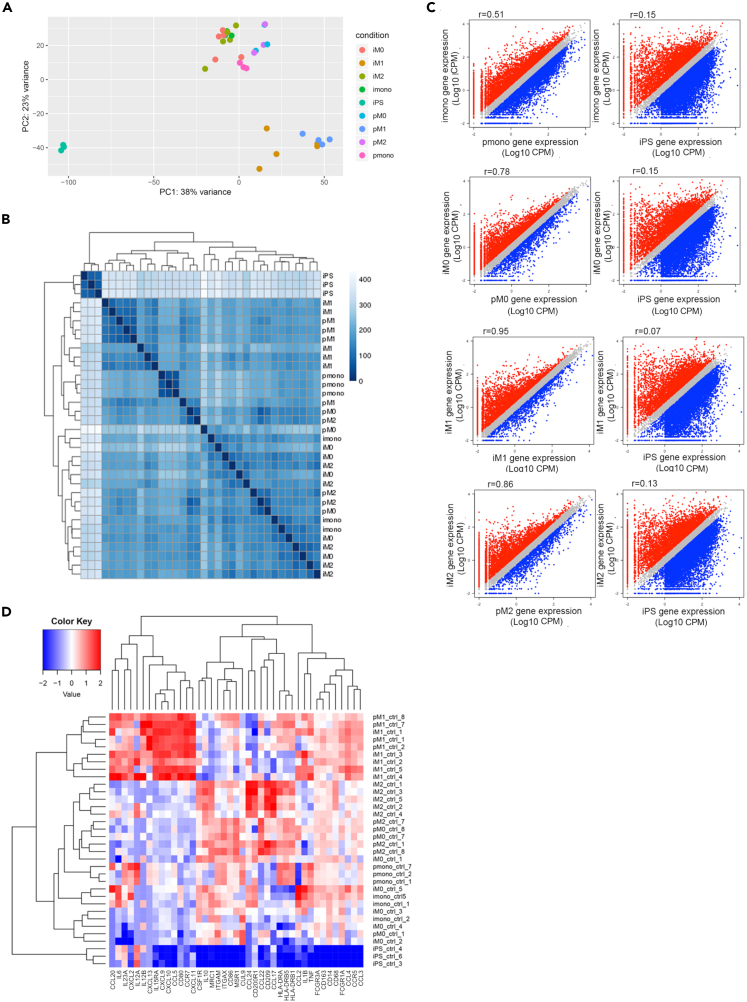
Figure 4.
Similarity of transcriptome between primary and iPSC-derived monocytes and macrophages
Analyses were performed based on RNA-seq data for both primary (p) and iMono and iMacrophages (M0, M1, and M2 according to sample polarization) samples.
(A) PCA analyses based on transcriptome for all healthy control RNA-seq samples.
(B) Hierarchical clustering based on transcriptome among all healthy control samples.
(C) Scatterplot of gene expression levels between primary monocytes and macrophages and iPSC-derived monocytes and macrophages. Comparison between iPSC-derived monocytes and macrophages and iPSCs were also shown as a control. Genes that are highly expressed in iPSC-derived samples are shown in red and genes that are highly expressed in primary monocytes and macrophages or iPSCs are shown in blue.
(D) Heatmap of genes associated with macrophages functions. Columns represent genes, and rows represent samples. See also Table S1.