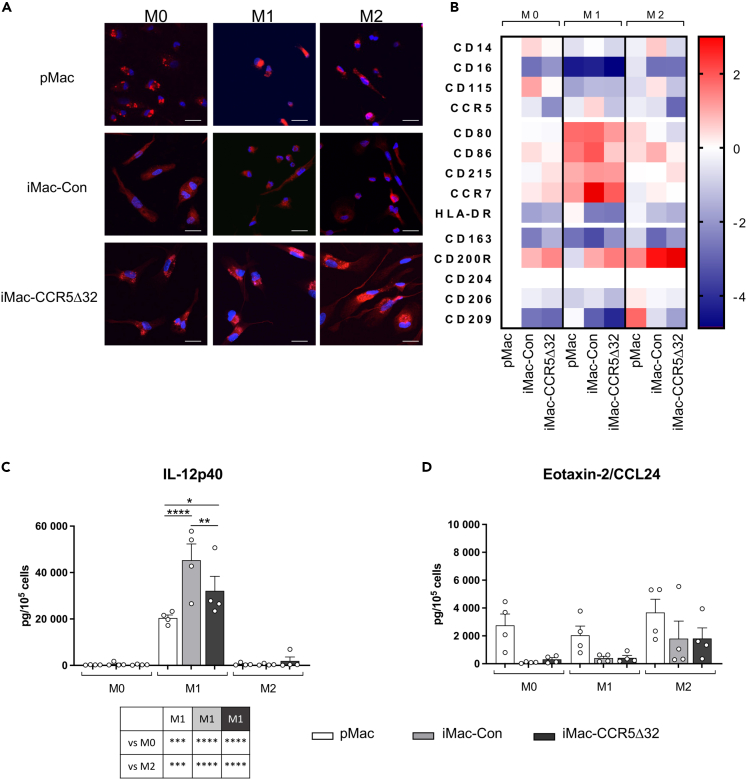
Figure 6.
Immunophenotype of primary and iPSC-derived macrophages
(A) Representative photographs of immunofluorescence CD68 staining observed in primary macrophages (pMac, first row, n = 3), iPSC-derived macrophages obtained from healthy controls (iMac-Con, second row, n = 3) and iMac obtained from individuals homozygous for CCR5Δ32 variant (iMac-CCR5Δ32, third row, n = 4) polarized in M0, M1, and M2 macrophages (First, second and third column, respectively). Cells were stained for CD68 (red) and with DAPI for nuclear staining (blue) (scale bar: 20 μm).
(B) Immunophenotype of pMac (n = 4), iMac-Con (n = 3) and iMac-CCR5Δ32 (n = 4) polarized in M0, M1, and M2 macrophages was evaluated by flow cytometry. Data are expressed as MFI. Data shown as Log2(Fold change) with all samples normalized on M0 pMac.
(C and D) Primary and iPSC-derived macrophage secretome profile. Expression levels of IL12p40 as M1 polarization marker (C) and Eotaxin-2/CCL24 as M2 polarization marker (D) were analyzed in primary (white bar; n = 4) and iPSC-derived M0, M1, and M2 macrophages obtained from both controls (light gray bar; n = 4) and CCR5Δ32 individuals (dark gray bar; n = 4). Data are expressed as pg/105 cells and are presented as mean ± SEM. Statistical significance was calculated using ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S3.