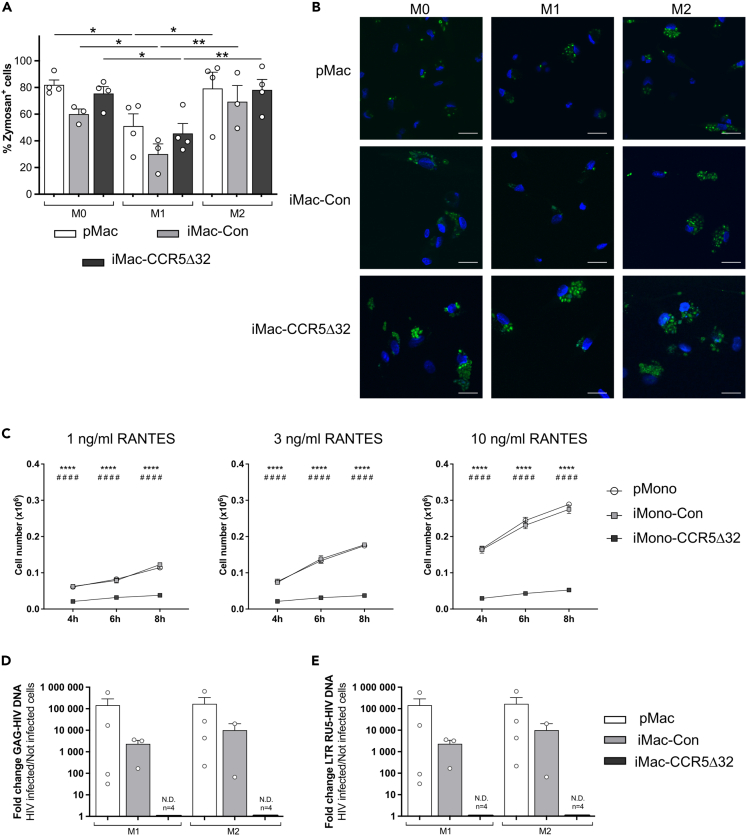
Figure 7.
Functionality of primary and iPSC-derived macrophages
Phagocytic ability of M0, M1 and M2 primary macrophages (pMac, white bar, n = 4) and iPSC-derived macrophages (iMac) obtained from both controls (iMac-Con, light gray bar; n = 3) and individuals homozygous for CCR5Δ32 variant (iMac-CCR5Δ32, dark gray bar; n = 4) was evaluated by using pHrodo Green Zymosan A Bioparticles and was assessed by flow cytometry and confocal microscopy.
(A) Macrophage phagocytic ability assessed by flow cytometry was shown as frequency of Zymosan+ cells.
(B) Representative photographs showing phagocytized zymosan particles (in green) in primary macrophages (pMac, first row, n = 3) and iMac obtained from healthy controls (iMac-Con, second row, n = 3) and iMac obtained from homozygous CCR5Δ32 individuals (iMac-CCR5Δ32, third row, n = 4) polarized in M0, M1, and M2 macrophages (First, second and third column, respectively). Nuclei were stained with DAPI (blue). Scale bar: 20 μm.
(C) The chemotactic ability of primary monocytes (pMono, white circle; n = 3) and iPSC-derived monocytes (iMono) obtained from healthy controls (iMono-Con, light gray square; n = 3) and from homozygous CCR5Δ32 individuals (iMono-CCR5Δ32, dark gray square; n = 3) to migrate in response to increasing the concentration of RANTES was assessed in vitro by a cell migration assay. The number of migrated cells was evaluated at different time points. ∗ iMono-Con vs. iMono-CCR5Δ32; # pMono-Con vs. iMono-CCR5Δ32.
(D and E) Macrophage infectability with HIV was assessed in M1 and M2 polarized primary and iPSC-derived macrophages obtained from healthy controls (iMac-Con) and from homozygous CCR5Δ32 individuals (iMac-CCR5Δ32) by evaluating through qPCR the presence of (D) specific HIV-1 DNA Gag and (E) specific HIV-1 DNA LTR RU5. Amplification of genomic GAPDH as a reference gene was used to control the amount of DNA in each sample. Data shown as mean ± SEM. Statistical significance was calculated using ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.