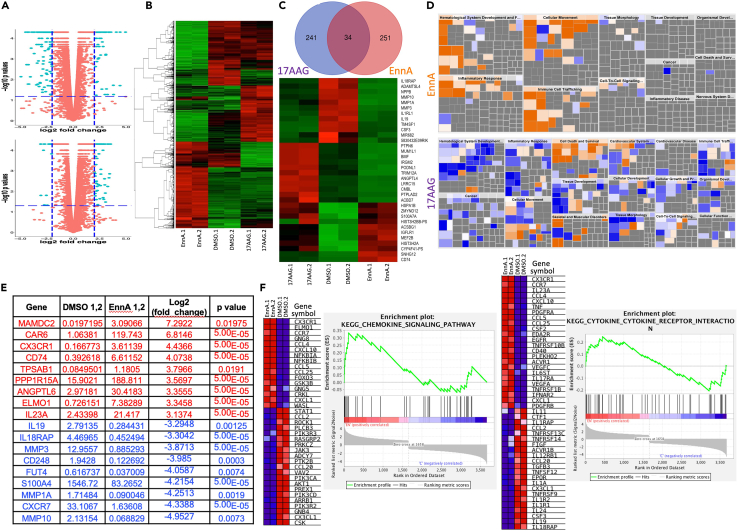
Figure 6.
EnnA modulates E0771 cell transcriptome to activate the immune system
(A) Volcano plots of bulk RNA-seq data from E0771 cells treated with EnnA, 17-AAG or DMSO as control. Plots were generated based on EnnA versus control (upper, +/− 2-fold change and p < 0.065, as shown in dashed blue lines) and 17-AAG versus control (bottom, +/− 2-fold change and p < 0.05, as shown in dashed blue lines), after the removal of low-expressing genes.
(B) Heatmap generated based on the intersection of 4852 genes with the normalized FPKM counts from duplicates of control, EnnA-treated, and 17-AAG-treated cells, after the removal of low-expressing genes.
(C) Venn Diagram (upper panel) indicates 251 genes at or exceeding the cutoff of 2 log2 fold changes in response to EnnA versus control-treated cells and 241 genes at or exceeding the cutoff of 1.5 log2 fold changes in response to 17-AAG versus control-treated cells, with an intersection of 34 genes, which were used to plot a heatmap across control, EnnA, and 17-AAG groups (lower panel).
(D) Predicted biological impact: Two filtered fold-change tables, EnnA versus control (upper, +/− 3-fold change and p < 0.05) and 17-AAG versus control (bottom, +/− 2-fold change and p < 0.05), were imported into IPA software to predict biological effects (increases (orange) or decreases (blue)) on diseases and cell biology processes.
(E) List of top genes affected by EnnA treatment. Red: upregulated. Blue: downregulated.
(F) Plots of Heatmap and Enrichment Score by GSEA on immune responses involving KEGG pathways. Heatmap and Enrichment Scores were plotted using the 3,668 genes at a cutoff of 1.5-fold changes into the GSEA analysis with the duplicates of EnnA versus control to produce the immune response-associated KEGG pathways containing chemokine signaling pathways and cytokine-cytokine receptor interaction. Data are represented as a mean ± S.E.M. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by one-way ANOVA or unpaired tailed Student’s t test. Data represent at least two independent studies.