Abstract
Autophagy is a multifaceted cellular process that not only maintains the homeostatic and adaptive responses of the brain but is also dynamically involved in the regulation of neural cell generation, maturation, and survival. Autophagy facilities the utilization of energy and the microenvironment for developing neural stem cells. Autophagy arbitrates structural and functional remodeling during the cell differentiation process. Autophagy also plays an indispensable role in the maintenance of stemness and homeostasis in neural stem cells during essential brain physiology and also in the instigation and progression of diseases. Only recently, studies have begun to shed light on autophagy regulation in glia (microglia, astrocyte, and oligodendrocyte) in the brain. Glial cells have attained relatively less consideration despite their unquestioned influence on various aspects of neural development, synaptic function, brain metabolism, cellular debris clearing, and restoration of damaged or injured tissues. Thus, this review composes pertinent information regarding the involvement of autophagy in neural stem cells and glial regulation and the role of this connexion in normal brain functions, neurodevelopmental disorders, and neurodegenerative diseases. This review will provide insight into establishing a concrete strategic approach for investigating pathological mechanisms and developing therapies for brain diseases.
Keywords: astrocyte, autophagy, glia, microglia, neural stem cells, neurodegenerative diseases, neurodevelopmental disorders, oligodendrocyte
Introduction
Autophagy means “self-eating” in Greek, and it is a dynamic catabolic pathway for lysosomal-mediated degradation of intracellular components and organelles (Filippelli et al., 2022; Lu et al., 2022). Autophagy is an evolutionarily conserved cellular quality control process (from yeast to mammalian cells) and it is essential for embryogenesis, survival of neonates, and adaptation to stress conditions. Autophagy can be induced by toxic, metabolic, oxidative, environmental, and inflammatory stresses, which subsequently target the protein aggregates, damaged organelles, and intracellular pathogens to lysosomes for clearance. This process recycles the degraded products to produce building materials and energy for cellular reconstruction. The autophagic events occur at both basal and induced levels, providing a routine constitutive turnover of cellular materials and organelles, in order to maintain tissue homeostasis. Three types of autophagy occur in mammalian cells, i.e., macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Chen et al., 2018; Lu et al., 2022). In all three types of autophagy, intracellular cargoes are delivered to lysosomes for degradation and recycling. Microautophagy implies the infoldings of the lysosomal membrane to catch a small portion of the cytosolic cargo. CMA employs cytosolic chaperones of heat shock cognate protein of 70 kDa to locate and seize the substrate. In contrast, in macroautophagy (hereafter autophagy), the cytoplasm cargoes and organelles are sequestered within double-membrane vesicles and then delivered to lysosomes for fusion and degradation.
The autophagic events are tightly regulated by the sequential activation of well-characterized autophagy-related genes and protein complexes (Figure 1). During the autophagy process, a double membrane cup-shaped phagophore initially forms to isolate the affected cytoplasm or organelles through a ULK1-Fip200-Atg13 complex (Ganley et al., 2009; Hosokawa et al., 2009). This phagophore then expands and encapsulates more cytoplasmic cargoes. Eventually, the phagophore closes and forms a complete vesicle known as an autophagosome with the LC3-PE conjugation system and Atg12-Atg5-Atg16l1 complex (Satoo et al., 2009; Romanov et al., 2012; Hollenstein and Kraft, 2020). The induction of autophagy can be initiated by stimuli that suppress the mammalian target of rapamycin complex 1 (mTORC1) pathway or stimulate the AMP-activated protein kinase (AMPK) energy-sensing pathway (Egan et al., 2011; Kim et al., 2011; González et al., 2020), key regulators for cell growth and energy metabolism. Prior to the delivery of its contents to lysosomes, autophagosomes can fuse with endosomes or multivesicular bodies and form a structure known as an amphisome (Fader et al., 2008). For the final steps, the autophagosome outer membranes tether with the lysosomal membrane to form an autolysosome for degradation of the materials it contains. The hydrolytic enzymes in the lysosome disintegrate the autophagosome contents and the resultant by-products, like free fatty acids, glycogen, and amino acids are used for further recycling as building materials and energy sources (Yim and Mizushima, 2020). A detailed description of autophagy can be found in some excellent reviews elsewhere (Lu et al., 2022; Zhen and Stenmark, 2023).
Figure 1.
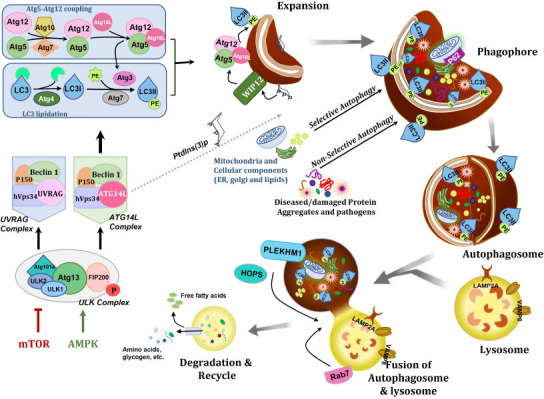
Signaling pathways and molecular mechanisms involved in autophagy.
Autophagy is a highly synchronized and complex self-degradative process that entails the cascades of various molecular complexes comprising autophagy-related genes and protein complexes. Autophagy can be triggered by AMPK stimulation leading to energy depletion and/or mTOR inhibition caused by nutrient starvation. Activation of autophagy initiates the formation of ULK complex, consists of ULK1/2, Atg13, Atg101a, and Fip200. Initiation of ULK complex instigates the PtdIn3K complexes, involving Atg14L, Beclin-1, VPS34, and VPS15. Atg14L complex functions in the nucleation and expansion of the phagophore via PtdIns (3) P generation. UVRAG complex contributes to the endo/lysosome system. Autophagosome maturation is regulated by the activation of the Atg5-Atg12 coupling system and lipidation of LC3. During Atg5-Atg12 coupling via Atg7 and Atg10, Atg12 is conjugated with Atg5 and later Atg16L to form the Atg5-Atg12-Atg6L complex. In LC3 lipidation, Atg4 cleaves LC3 at the C terminus and generates LC3-I. Then, via Atg7 and Atg3, LC3-I is coupled to PE. In the expansion phase, with WIPI2, Atg5-Atg12-Atg6L complex localizes on the surface of a phagophore, and further prompts LC3 lipidation and Atg3 stimulation. For degradation, non-selectively/selectively damaged/diseased protein (Aβ, tau etc.) aggregates (aggrephagy), pathogens (xenopahgy), and specific cellular components marked with adaptor protein like p62/SQSTM1 (sequestosome 1) and lipid droplets (lipophagy), are confined within the double membrane phagophore. Then, the autophagosome completes phagophore membrane expansion and closure through LC3 support. Later, lysosomes with protein Rab7 fuse with the fully formed autophagosome to degrade the intra-lysosomal components. Successively, autophagy targets get degraded and their building blocks, such as free fatty acids and amino acids, are recovered back into the cytosol. Created with Microsoft PowerPoint. AMPK: AMP-activated protein kinase; Fip200: focal adhesion kinase family interacting protein of 200 kDa; HOPS: homotypic fusion and protein sorting; LAMP2: lysosomal associated membrane protein 2; LC3: microtubule-associated protein 1A/1B-light chain 3; mTOR: mammalian target of rapamycin; PE: phosphatidylethanolamine; PLEKHM1: pleckstrin homology domain-containing family M member 1; Rab7: Ras-related protein 7; ULK: Unc-51 like kinase; UVRAG: UV radiation resistance-associated gene protein; VPS15: vacuolar protein sorting 15; VPS34: vacuolar protein sorting 34; WIPI2: WD repeat domain, phosphoinositide interacting 2.
Neural stem cells (NSCs) are multipotent self-renewing cells residing in neurogenic niches that are responsible for producing new neurons and glia in embryonic and adult brains (Obernier and Alvarez-Buylla, 2019). With adaptive response functions for maintaining brain homeostasis, NSCs are actively involved in neurodevelopment, tissue repair, and brain regeneration (Obernier and Alvarez-Buylla, 2019; Rodríguez-Bodero and Encinas-Pérez, 2022). NSCs are long-lived cells throughout the lifespan of an organism and quality control systems are critical for their health and proper functions. Previous studies indicate that autophagy is involved in the generation of new neurons (neurogenesis), maintenance, and self-renewal of NSCs (Doetsch, 2003; Liu et al., 2021). There is an increase in the expression of the autophagy genes Atg7, Beclin1, Ambra1, and LC3 in the olfactory bulb during the initial period of neuronal differentiation in mouse embryos. It is suggested that autophagy promotes neurogenesis by providing energy to NSCs to facilitate cytoskeleton remodeling during differentiation (Vázquez et al., 2012; Ordureau et al., 2021). Furthermore, autophagy in NSCs not only supports the sustainability of building materials but also preserves homeostasis by preventing the growth of pathological and toxic protein aggregates and delivering damaged or unnecessary cellular components to lysosomes for degradation. By maintaining a balance between synthesis and degradation in the maintenance, differentiation, and growth of NSCs, autophagy retains healthy brain development and functions. Defects of autophagy may also result in NSCs instability and aberrant differentiation. These processes may underlie neurodevelopmental disorders such as autism spectrum disorder (ASD) (Poultney et al., 2013), childhood ataxia (Kim et al., 2016), primary microcephaly (Kadir et al., 2016), and genetic leukoencephalopathies (Deng et al., 2021). It is also possible that the multi-faceted roles of autophagy may affect NSCs maintenance and differentiation in ischemic stroke (Xu et al., 2022). Prolonged autophagy or upregulation of autophagy can result in autophagic cell death (Klionsky et al., 2016; Jung et al., 2020a). A recent study has shown that under chronic stress, NSCs undergo autophagic cell death (Jung et al., 2020b). An NSC-specific Atg7 conditional knockout attenuated the decay of adult hippocampal neurogenesis and mitigated cognitive deficits by reducing the autophagy flux in stressed animals (Jung et al., 2020b). This review will discuss the multi-faceted roles of autophagy in postnatal NSCs for the progression of neurodevelopmental disorders and ischemic brain injuries.
Glia, including astrocytes, microglia, oligodendrocytes, and ependymal cells, are the majority of the cell population in the brain (Sun et al., 2022). In embryonic development, NSCs are initially multipotent but are gradually restricted to neuronal or glial destinations. Neural differentiation ensues in a stereotypical process whereby neurons are generated first, followed by glia (mainly astroglia and oligodendrocytes), which differentiate from glial progenitors after completion (mostly) of neurogenesis. Microglia are myeloid cells of embryonic hematopoietic origin that migrate to the brain during the early stages of brain development. Glial function to support neurons during development and throughout their lifetime. They are key players in maintaining brain functions and homeostasis through their plethora of physiological functions ranging from neurotransmission support, energy supply, blood flow regulation, immune surveillance, and more (Henn et al., 2022). Glia participate in the pathophysiology of various neurodegenerative diseases, where they respond as a double-edged sword, depending on the severity of injuries and the nature of the diseases (Henn et al., 2022). Glia may accelerate brain repair through phagocytosis, metabolic support, and secretion of growth factors, but they can also devastate by generating uncontrollable neuroinflammatory responses (Vainchtein and Molofsky, 2020; Henn et al., 2022). Considering the involvement of autophagy in both adaptive and innate immunity (Levine and Deretic, 2007; Metur and Klionsky, 2021), it is not surprising to find the critical functions of autophagy in glia functioning to regulate inflammatory signaling, pathogen elimination, maintenance of immune modulators, and modification of antigen presentation in various neurodegenerative diseases. Nevertheless, the current research has just begun to unveil the involvement of autophagy in the regulation of glia and their allied effect on neurodegenerative diseases.
Contemplating the similar origin of glia with NSC and the involvement of autophagy, extensive studies are needed to better understand the connexion between these essential brain processes. The information will aid in determining mechanisms and targets to treat developing, adult, and aging brains. This present review compiles current information on autophagy in the regulation of NSC and glial functions and its implication for neurodevelopmental disorders and neurodegenerative diseases.
Autophagy in Neural Stem Cells
Roles of autophagy in NSC regulation
NSCs give rise to a wide range of new neurons and glia in the embryonic nervous system. In the adult brain, continuous postnatal NSCs are primarily confined to two regions: the subventricular zone (SVZ) of the lateral ventricle and the subgranular zone (SGZ) of the hippocampus (Yazdankhah et al., 2014; Obernier and Alvarez-Buylla, 2019; Llorente et al., 2022). NSCs can generate a huge number of neurons and glia in embryonic and postnatal brain development. Postnatal NSCs in SGZ and SVZ have a lifelong neurogenesis capability, which has a great impact on learning, memory, behavior, and restoring the cell after damage (Mirzadeh et al., 2008; Llorente et al., 2022). Autophagy enforces a protective effect in the production and differentiation of NSCs by the removal of damaged organelles and proteins. Consequently, autophagy takes part in brain growth, development, and pruning of unused synapses from newborn neurons, and controls the subsistence or mortality of the NSCs under unfavorable conditions.
Proliferation and differentiation of NSCs are essential elements for their self-renewal and damage repair processes. Our work to produce a conditional knockout (cKO) of an essential autophagy gene Fip200 (also known as RB1 inducible coiled-coil 1, Rb1cc1) in mice for the first time revealed the essential function of autophagy proteins in postnatal NSC maintenance and differentiation (Wang et al., 2013). Deletion of Fip200 in young adult mice (1-month-old) leads to a reduction in the adult NSCs pool and switches neurogenesis to glial differentiation into astrocytes. These protective effects of Fip200 are through the suppression of ROS-mediated (reactive oxygen species) p53 activation in postnatal NSCs. In follow-up studies to knock out other autophagy genes (Atg5, Atg7 and Atg16l1), our work reveals the essential roles of autophagy receptor p62/SQSTM1 aggregations in NSCs for the generation of ROS by sequestering SOD1 in nuclei (Wang et al., 2016). We observed fewer p62/SQSTM1 aggregations in Atg5 cKO and Atg16l1 cKO NSCs and do not find NSCs defects, which suggest autophagy gene-specific mechanisms for postnatal NSCs development. Similarly, autophagy inhibition through Beclin-1 deletion or class III PI3K inhibitor 3-methyladenine results in a substantial repression of NSCs maintenance and differentiation (Yazdankhah et al., 2014). Activating molecule in beclin1-regulated autophagy 1 (Ambra1) cKO in mouse embryos causes neural tube defects related to distorted cell proliferation, accretion of ubiquitinated proteins, and excessive cell death (Fimia et al., 2007). Similarly, a supportive role of autophagic genes such as Becn1, Atg7, LC3, and Ambra1 is observed in the neurogenesis of mouse embryo olfactory bulb (Vázquez et al., 2012). Besides these studies of core autophagy genes, another autophagy-related gene Eva1a (also known as transmembrane protein 166, TMEM166) has been suggested in controlling the differentiation and self-renewal of NSCs through activation of PI3K-AKT axis and mTOR in the embryonic brain (Li et al., 2016). Moreover, the inactivation of Forkhead box O (FoxO) transcription factors (FoxO1, FoxO3, and FoxO4) leads to defective differentiation and self-renewal of NSCs (Paik et al., 2009). Recent evidence also confirms a critical role of FoxOs in brain development via the regulation of autophagy in NSCs (Schäffner et al., 2018). Interestingly, a recent study of NSC aging reveals that enhancing autophagy function in a Beclin-1 knockin mutation mouse (Fernández et al., 2018) reduces the decline rate of NSC loss and restores neurogenesis in SVZ of aged knockin animals (Wang et al., 2022). Based on the aforementioned evidence for the physiological functions of autophagy in NSCs with involvement in proliferation, differentiation, and self-renewal potential of NSCs, it is necessary to warrant more research to reveal autophagy targets and their precise mechanisms in NSC regulation at different development/aging stages. Such information would provide better targets and/or therapeutic strategies for the treatment of neurodevelopment disorders.
Autophagy in NSCs of brain disorders
Multipotent embryonic NSCs differentiate into neurons, astrocytes, and oligodendrocytes to form the central nervous system (CNS) in mammals. In the CNS, NSCs start differentiation and migrate within two specific brain regions of the telencephalon, i.e., the SVZ/rostral migratory stream/olfactory bulb path and the SGZ/GZ path, both of which are vital for the physiological processes of food chasing, mating, learning, memory formation, and so on. Thus, both embryonic and postnatal NSCs play essential roles in the formation of neural circuits, maintenance of neuroplasticity, brain repair, aging, and cognition. Therefore, it is not surprising that dysfunctional autophagy in NSCs is involved in the initiation and progression of various neurological disorders, ranging from neurodevelopmental disorders in young individuals to brain injury of ischemic stroke in adults or aged ones.
Autophagy in NSCs of neurodevelopmental disorders
Neurodevelopmental disorders are characterized as multifaceted groups of mental abnormalities with subsequent cognitive deficits and behavioral impairment. Throughout postnatal brain development, autophagy critically maintains neural plasticity and homeostasis by removing toxic aggregates, proteins, and damaged organelles. The absence of autophagy in NSCs and maturing neurons leads to axon dystrophy, aberrant synaptic transmission, deposition of ubiquitinated proteins, and degeneration that further leads to neurodevelopmental disorders (Levine and Kroemer, 2019). A pivotal link of autophagy alteration to neurodevelopmental disorders is highlighted by a recent human study that shows that recessive variants of Atg7 lead to complicated neurodevelopmental disorders with malformations of the corpus callosum and cerebellum (Collier et al., 2021). In NSCs, it is postulated that altered autophagy, impaired autosomal-lysosomal processes, and altered proteasome functions make NSCs more susceptible to toxic metabolite accumulation. Thus, restoring normal autophagy function is a potential therapeutic approach. We have summarized the involvement of autophagy in various neurodevelopmental disorders in Table 1.
Table 1.
Autophagy-related gene mutations causing neuro-disorders in development
| Gene symbol | Functions in autophagy | Related disease(s) | Clinical features |
|---|---|---|---|
| ATG5 | A core autophagy gene involved in autophagosome elongation, maturation, and closure; via conjugation with ATG12, acts as an E3-like enzyme required for lipidation of ATG8 family proteins and their association to the vesicle membranes. | Autosomal recessive spinocerebellar ataxia 25 (SCAR25) | Delayed psychomotor development, truncal ataxia, dysmetria, nystagmus, low IQ, cerebellar hypoplasia |
| WDR45 (WIPI4) | Autophagosome-lysosome fusion, autophagosome formation and elongation. | Beta-propeller protein-associated neurodegeneration (BPAN) | Cognitive decline, dementia, dystonia, parkinsonism, optic nerve atrophy, global developmental delay, intellectual disability, psychomotor retardation, febrile seizures, autistic features |
| WDR45b (WIPI3) | Autophagosome-lysosome fusion, autophagosome formation and elongation. | Neurodevelopmental disorder with spastic quadriplegia and brain abnormalities with or without seizures (NEDSBAS) | Absent speech, spastic paraplegia, intellectual disability, profound inability to walk, cerebral hypoplasia, thin corpus callosum, enlarged ventricles, delayed psychomotor development, profound reduced white matter volume, refractory seizure (in most patients), absent communication, thinning of the cortex |
| EPG5 | Autophagosome-lysosome fusion. | Vici Syndrome | Cataracts, hypopigmentation, muscle and neurogenic anomalies, immunodeficiency, corpus callosum agenesis, epilepsy, cardiomyopathy, microcephaly, loss of learned skills |
| TECPR2 | Autophagosome-lysosome fusion, Autophagosome formation. | Spastic paraplegia type 49 | Spasticity, pyramidal weakness, dysplastic corpus callosum |
| SPG15 (KIAA1840) | Autophagosome maturation, lysosome biogenesis | Hereditary spastic paraplegia, autosomal recessive axonal neuropathy, Charcot–Marie–Tooth disease | Spasticity, pyramidal weakness, corpus callosum affection, ataxia (less common) |
| SNX14 | Autophagosome-lysosome fusion | Autosomal recessive spinocerebellar ataxia 20 (SCAR20), aceruloplasminemia | Purkinje cell loss, mental retardation, seizures, ataxia, cerebellum atrophy, hearing loss, relative macrocephaly |
| TBCK | mTOR activator, regulator of small GTPases of the Rab-family. | TBCK encephaloneuropathy | Leukoencephalopathy, coarse face, intellectual disability, seizures, hypotonia, neuronopathy |
| PTEN | antagonizes the phosphatidylinositol 3-kinase class I/AKT/mTOR pathway | Lhermitte-Duclos disease | Autism, macrocephaly, ataxia, seizures, gangliocytomas in the cerebellum |
| NF1 | mTOR inhibition | Neurofibromatosis | Autism, epilepsy (10%), café-au-lait spots, malignant and benign tumors |
| Fmr1 | mTOR inhibition | Fragile X syndrome | Intellectual disability, autism, seizures, hypersensitivity, attention deficit, hyperactivity, growth, and craniofacial abnormalities |
| TSC1/2 complex | mTOR inhibition | Tuberous sclerosis complex | Non-malignant tumors, epilepsy, autism, TAND |
AKT: Ak strain transforming; ATG5: autophagy-related gene 5; ATG8: autophagy-related gene 8; ATG12: autophagy-related gene 12; EPG5: ectopic P-granules 5 autophagy tethering factor; Fmr1: fragile X messenger ribonucleoprotein 1; GTPases: guanosine triphosphatases; IQ: intelligent quotient; LC3II: microtubule-associated protein light chain 3II; mTOR: mammalian target of rapamycin; NF1: neurofibromatosis 1; PTEN: phosphatase and tensin homolog; SCAR25: spinocerebellar ataxia, autosomal recessive 25; SNX14: sorting nexin 14; SPG15: spastic paraplegia 15; TAND: tuberous sclerosis associated neuropsychiatric disorders; TBCK: homozygous TBC1 domain-containing kinase; TECPR2: tectonin beta-propeller repeat containing 2; TSC1/2: tuberous sclerosis complex1/2; WDR45 (WIPI-4): WD-repeat β-propeller protein 45; WDR45b (WIPI3): WD-repeat β-propeller protein 45b.
ASD
ASD is one of the most prevalent multi-factorial neurodevelopmental disorders caused by the combination of genetic and environmental factors. ASD is characterized by repetitive and restricted behavior, altered social interactions with verbal and non-verbal communication (Sandin et al., 2017; Deng et al., 2021). Concomitantly, ASD patients also show other comorbidities with intellectual disability, sensory processing defects, and motor deficits (Lazaro et al., 2015; Balasco et al., 2020; Al-Beltagi, 2021). Pathway and enrichment analyses of autophagic genes of MAP1LC3B, MAP1LC3A, GABARAPL2, GABARAP, and GABARAPL1 (mammalian orthologs to yeast Atg8) altered by deletion in ASD cases in both human and animal studies were found (Poultney et al., 2013; Hui et al., 2019). Other ASD-related genes, such as TSC1, TSC2, mTOR, and PTEN, are all closely related to the regulation of autophagy in NSC development and differentiation (Wang et al., 2019a; Deng et al., 2021). Autophagy has been implicated in synaptic function during the maturation of neurons in ASD patients. Tang et al. (2014) indicate that autophagy is impaired in ASD, while the pruning of excess synapses through neuronal autophagy is altered in developing ASD neurons. These studies indicate that autophagy dysfunction in NSCs development might be a central mechanism for the pathogenesis of ASD. Under certain physiological and pathological states, cargo-specific removal of organelles by selective autophagy is crucial to sustain the viability and stability of the brain milieu. One-way neutral lipid storage in lipid droplets is mobilized by autophagy to release free fatty acids. This process is known as lipophagy (a form of selective autophagy) (Singh et al., 2009) and it is regulated by protein receptors on the surface of the lipid droplets (Singh and Cuervo, 2012). After lipid droplets sequestration in a double-walled membrane vesicle, they are transported to lysosomes and degraded by liposomal acidic lipase (Figure 1). In the brain, NSCs express genes for fatty acid synthesis and fatty acid oxidation to generate adenosine triphosphate, and it was observed that inactivation of lipid biogenesis could cause alteration in the NSC activity that could lead to dysfunctional maintenance and differentiation. Additionally, a study on TSC-deficient animal models has shown that lipophagy inhibition through genetic knockout of Fip200 and pharmacological inhibitors could help restore NSC functions and impede the progression of benign brain tumors (Wang et al., 2019a). In the future, interventions of selective autophagy instead of bulk autophagy might be more effective and specific for the treatment of some ASD patients.
Childhood ataxia
Childhood ataxia is a rare disease affecting any level of the nervous system and comprised of diminished coordination of balance and movement as well as cognitive disability and behavioral developmental delay in children (Kim et al., 2016; Pavone et al., 2017). A homozygous missense mutation of Atg5 (E122D) that diminishes autophagic activity causes congenital ataxia (Kim et al., 2016; Kawabata and Yoshimori, 2020).
Primary microcephaly
Primary microcephaly is a heterogeneous congenital neurodevelopmental disorder as a result of diminished production of neurons during brain development, leading to reduced head circumference and brain volume (Jean et al., 2020). The pathogenic mutations in the human WDFY3 gene that encodes an autophagy scaffold protein are identified in primary microcephaly (Kadir et al., 2016; Le Duc et al., 2019). Ablation of WDFY3 in the brain also reduces developmental axonal connectivity and impairs the formation of the major forebrain commissures in a mouse model (Dragich et al., 2016).
Genetic leukoencephalopathies
Genetic leukoencephalopathies are a set of rare genetic heterogeneous disorders that lead to progressive degeneration of white matter in the CNS. The predominant symptoms in these disorders are motor impairment, ataxia, and altered cognitive development (Sarret, 2020). Mutations in VPS11, a core component of class C vacuole/endosome tethering proteins, are observed in autosomal recessive leukoencephalopathy patients (Lee et al., 2013; Vanderver, 2016). VPS11 is involved in the lysosome-degradation and membrane trafficking during autophagy in human cells (Byrne et al., 2016). The SNF8 mutation (C846G) triggers abnormal ubiquitination and accelerated turnover of VPS11 protein, suggesting the regulation of the autophagy degradation pathway in developing leukoencephalopathies (Zatyka et al., 2020).
Autophagy in NSCs of ischemic stroke
Ischemic stroke is considered to be the leading cause of disability and preventable mortality worldwide. During ischemic stroke, reduced blood flow to the brain leads to cerebral vascular occlusion and stimulates a severe inflammatory rush comprised of toxic mediators, harmful oxidative radicals, and inflammatory macrophage infiltration that further exacerbates brain tissue damage and cellular death. Inflammation works as a double-edged sword during ischemic injury as it also protects the brain by tissue remodeling and repair (Sakai and Shichita, 2019). Studies have shown that stimulation of the autophagy gene Beclin-1 reduces the cortical inflammation initiated by ischemic brain injury in rats (Zhou et al., 2011). Diminished regenerative ability is also a contributor to ischemic stroke pathogenesis. NSCs have been deemed as an effective regenerative and neuroprotective therapy against ischemic stroke. In the mammalian brain, ischemic stroke initiates the genesis of cells in the SVZ and SGZ via NSCs and these newly generated cells migrate toward the ischemic lesion boundary and support the recovery of damaged brain tissue (Zhao et al., 2018). Alterations of autophagy in NSC might hamper the recovery of the ischemic brain. Autophagy not only regulates NSCs healthy homeostasis but also ensures NSC programmed death if stimulated by prolonged trauma (Jung et al., 2020). Studies have shown that pharmacologically activated autophagy in the ischemic brain reduces the capability of NSCs and delays recovery (Wang et al., 2019b; Jung et al., 2020). Stimulation of autophagy in NSCs helps by inhibiting the ischemic brain damage to some extent, though excessive autophagy can aggravate the damage and lead to cell death. The dual aspect of autophagy in NSC during ischemia is still under investigation.
Therapeutic implication of autophagy modulation in neurodevelopmental disorders
Autophagy has a wide range of impacts on various physiological and pathological conditions. Thus, modulation of autophagy genes and related pathways may be considered as potential therapeutic targets for the treatment of neurodevelopment diseases. During spinocerebellar ataxia, modulation in autophagy supports the removal of abnormal ataxin-3 protein (Menzies et al., 2010). Similarly, trehalose, an mTOR-independent autophagy inducer and its analogs melibiose and lactulose, was used to regulate the ataxin-3 protein following spinocerebellar ataxia (Lin et al., 2016; Watchson et al., 2023). Considering the autophagic function of clearing the excessive intracellular protein accumulation, pharmacological modulation of the autophagic-lysosomal pathway by blocking mTORC1 was observed as a critical neuroprotective strategy for proteinopathies (Thellung et al., 2018). Apparently, pharmacological therapy using rapamycin has been moderately successful in restoring behavioral and cognitive symptoms in various neurodevelopment disorders, like ASD (Wang et al., 2023) and tuberous sclerosis complex (Ehninger et al., 2008; Sato et al., 2012). Nevertheless, to the best of our knowledge, direct evidence of the prospective effect of autophagy modulation for the maintenance and regulation of NSC for neurodevelopment disease is still scanty. The neurodevelopment studies conducted on animal and cell line models have predicted the involvement of autophagy in the survival of NSC, neurons, and glia. Hence, it is reasonable and enticing to speculate that modulation of autophagy genes and activation of autophagy pathways in postnatal NSCs would develop promising therapeutic strategies against neurodevelopmental diseases.
Autophagy in Glia
The contribution of autophagy to glia regulation is still unexplored, although glia encompasses 50% or more of the brain cell population. Glia are crucial regulators of neuronal homeostasis and brain functions. They directly impact brain development, synaptic function, metabolism, and restoration after damage or injury. Some recent studies propose that autophagy plays an essential role in gliogenesis (Belgrad et al., 2020; Deng et al., 2021). Existing studies on neurodegeneration, nerve injuries, and aging assessing the involvement of autophagy are largely focused on neurons. It is plausible that any alteration in glia generation and/or differentiation might lead to structural and functional brain abnormality. Scant evidence shows the contribution of autophagy to the regulation of glial function under normal and pathological conditions (Jin et al., 2018; Chandrasekaran et al., 2021). In glia, consideration of the therapeutics of autophagy modulation is quite limited due to partial information about the autophagy genes and/or pathways involved in the development and neurodegenerative diseases. Moreover, understanding is lacking about the specificity of pharmacological inhibitors/enhancers for autophagy functions in glia. In this section, we will review the functions and regulation by autophagy of major glial cell types in the brain (Figure 2).
Figure 2.
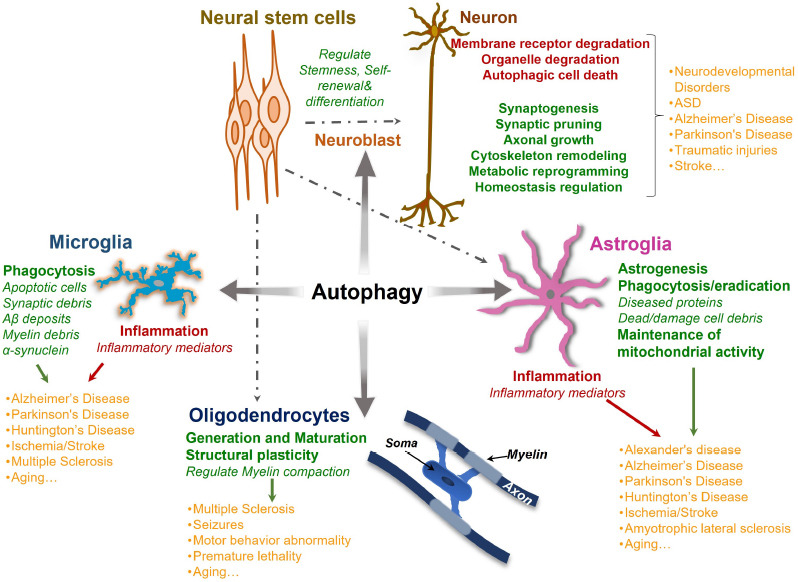
Autophagy connexion with brain cells in health and diseases.
Autophagy plays a crucial role in the development and maintenance of the brain and its functions. By promoting neural stem cell differentiation and self-renewal, autophagy takes part in structural and metabolic reprogramming and quality control functions during brain development. Autophagy not only ensures healthy brain development and processing but also protects the brain from various neurodevelopmental and neurodegenerative disorders in later life. Similarly, autophagic regulation of glia (microglia, astroglia, and oligodendroglia) function and structure helps in maintaining the central nervous system milieu. During neurodegenerative disease, autophagy aids the glia in clearing the damaged and diseased cellular components and regulating inflammation mediators. The aberrant autophagic regulation of glial functions can stimulate or amplify the development of various neurological disorders. Created with Microsoft PowerPoint. Aβ: Amyloid-beta; ASD: autism spectrum disorder.
Autophagic glia in healthy brain
Microglia
Microglia, the brain resident macrophages, attain phagocytic attributes to remove damaged/dead cells or synapse debris, orchestrate brain surveillance, respond to CNS damage, and release various inflammatory mediators (Borst et al., 2021; Butler et al., 2021). Autophagy in myeloid lineage regulates both adaptive and innate immune responses (Jiang et al., 2019; Pradel et al., 2020), phagocytosis (Wu and Lu, 2020; Li et al., 2021), and affects the consequences of inflammatory responses (Cho et al., 2020; Minchev et al., 2022) and antigen presentation (Germic et al., 2019; Münz, 2021). Nevertheless, few studies have assessed the involvement of autophagy in microglia regulation and the influence of autophagy dysregulation on the physiological functions of microglia. Atg7 deletion in microglia impairs synaptic connectivity, alters synaptosome degradation, and increases dendritic spines and synaptic markers (Kim et al., 2017). Microglia-specific ablation of Atg7, but not Ulk1, leads to the accretion of phagocytosed myelin and promotes the pathogenesis of multiple sclerosis-like disease (Berglund et al., 2020). A study has shown that Atg7 deletion in microglia impairs the homeostasis of mature oligodendrocytes and enhances susceptibility to acute and lethal generalized seizures (Alam et al., 2021). In vitro, BV2 cells increase neurotoxicity after Atg5 deletion but reduce it after treatment with mTORC1 inhibitor rapamycin or AKT inhibitor perifosine in TNF-α stimulated conditions (Jin et al., 2018). These data suggest Atg5 regulates microglia polarization during neuroinflammation (Jin et al., 2018). Collectively, microglia autophagy includes the transportation of aggregated proteins, injured organelles, and other cellular metabolite accumulation into lysosomes. Thus, regular autophagy in microglia not only provides plausible essential support to the homeostasis of microglia, but also enhances microglia defence mechanism during stress and disease conditions.
Astrocytes
Astrocytes are a major glial population in the CNS. They are involved in the establishment of neuronal synaptic networks, generation of neurotrophic factors, regulation of neurogenesis, recycling of neurotransmitters, supply for nutrition, and detoxification to maintain brain homeostasis (Haim et al., 2015; Sofroniew, 2020). Astrocytes also take part in brain immune regulation by releasing inflammatory molecules during neurodegenerative disease (Sofroniew and Vinters, 2010). Information regarding autophagic mechanisms that regulate astroglia functions is still scanty, although recent studies reveal autophagy’s critical roles in synchronizing astroglia functions. It has been shown that autophagy can decrease protein accumulation and enhance cell viability in in vitro cultured astrocytes (Jänen et al., 2010). In Alexander’s disease, autophagy degrades the mutant glial fibrillary acidic protein in astrocytes to prevent its accumulation (Tang et al., 2008). In late embryonic cortex development, autophagy is engaged in astrocyte differentiation. Atg5 deletion in mice diminishes the differentiation of NSCs into astrocytes via inhibiting the SOCS2-JAK-STAT (suppressors of cytokine signaling 2-janus kinase-signal transducer and activator of transcription) signaling pathway, while overexpression of Atg5 causes excessive astrocyte differentiation (Wang et al., 2014). This autophagy regulation is studied in the adult hippocampus since inhibition of autophagy leads to reduced astrocyte differentiation (Ha et al., 2019). Neuroinflammation leads to the fragmentation of mitochondria in astrocytes, which are engulfed by autophagosomes to support the restoration of tubular mitochondria. The absence of autophagy enhances ROS production from the fragmented mitochondria and reduces cell viability (Motori et al., 2013). Overall, astrocyte autophagy is less studied compared to autophagy in neurons and microglia. Regarding the evidence so far, astrocyte autophagy has the potential to influence the pathology of neurodegenerative diseases.
Oligodendrocytes
Oligodendrocytes endure vigorous structural and functional changes, comprising massive protein generation, lipid production, and membrane compaction. These processes require autophagy in development, neurodegenerative disease, and injury. Atg5 is essential for oligodendrocyte development through enhanced autophagic flux, increased autophagosome puncta, and elevated levels of LC3-II in the distal processes of oligodendrocytes during differentiation (Bankston et al., 2019). Additionally, Atg5 reaches the optimum cytoplasm level and maintains myelin compaction in oligodendrocytes (Bankston et al., 2019). Autophagy-deficient animals show enhanced populations of mature oligodendrocytes and increased levels of myelin protein which result in severe seizures (Alam et al., 2021). A recent study shows that genetic ablation of macroautophagy in oligodendrocytes caused an age-dependent surge in myelin thickness and abnormal myelin structures, accompanied by neurodegeneration, behavioral abnormalities, and premature lethality (Aber et al., 2022).
Summary
The information regarding autophagy involvement in gliogenesis and differentiation is still scanty. Previous studies have provided clues to understand autophagy in the regulation of glial development and function. However, to explore this, a glia-specific (e.g., microglia, astrocytes, and oligodendrocytes) deficiency of autophagy genes might be required to clarify how glia autophagy influences brain development and function under both health and disease conditions. Moreover, a better understanding of the mechanisms by which the autophagy influences gliogenesis and differentiation may require novel ways to modulate the endogenous glial progenitor cells within the brain for regenerative therapies.
Autophagy in glia of aging and diseased brain
Roles of glial autophagy in aged brain
Aging is a complicated biological event that can partially be explained by the accretion of molecular and cellular damages to brain cells over time. The inability to repair such damages in the brain may lead to subsequent failure of basic physiological functions, such as cognition, motor functions, and sensory functions that decrease the quality of life. During aging both microglia and astrocytes undergo molecular and morphological changes. A low-grade microglia-driven chronic inflammation, called “inflamm-aging” was reported in the aging brain. The stimuli that instigate and retain such inflammation during aging occur due to the accumulation of undigested cell waste from damaged organelles and cytoplasm (Franceschi et al., 2017) indicating an impaired lysosomal clearance process, like autophagy and/or phagocytosis. Deficiency of Atg7 is correlated to the appearance of age-associated inflammatory responses in peripheral macrophages. Atg7 deficiency showed an elevated level of inflammatory cytokines and severe inflammasome reaction (Stranks et al., 2015). Notably, autophagy controls the activation of inflammasomes (Takahama et al., 2018). Further studies have shown that alterations in selective autophagic pathways, like, mitophagy and pexophagy, are implicated in age-associated brain disorders. Diot et al. (2016) suggest that a decrease in mitophagy might be a crucial factor of human aging. Considering the involvement of autophagy in removing oxidative stress-led damages to organelles and macromolecules in the aging brain, it is a vital regulatory mechanism in maintaining homeostasis (Vellai et al., 2009). These studies suggest an emerging view of the involvement of glial autophagy in the aging brain. There are numerous studies available for the autophagic events in the aging brain, though few discuss autophagy in glia. Further studies are required to show the significance of glial autophagy during aging. Moreover, the lifestyle, dietary adjustments, and pharmacological interventions to improve autophagy could aid in reducing or suppressing the risk of progressing age-associated neurodegenerative disorders.
Roles of glial autophagy in neurodegenerative diseases
Aging is the most relevant negative factor for neurodegenerative disease. In this section, we will review the functions and regulation of autophagy in major glial cell types with different neurodegenerative diseases. We focus on the role of autophagy in microglia and astrocytes for their involvement in Alzheimer’s disease (AD) and Parkinson’s disease (PD; Figure 3).
Figure 3.
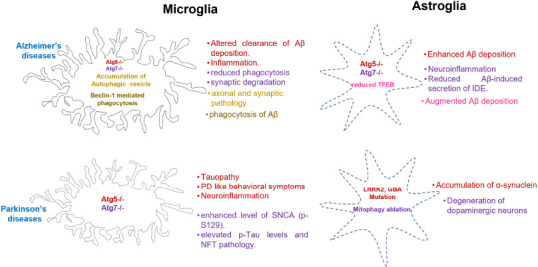
Autophagic gene and pathway abnormalities in microglia and astrocytes affecting pathophysiology of AD and PD.
Diagram illustrating various autophagy genes and pathways in microglia and astrocyte aggravating the pathology of AD and PD brain. Text of the same color indicating the autophagic gene or pathway with causative symptoms. Created with Microsoft PowerPoint. AD: Alzheimer’s disease; Aβ: amyloid-beta; GBA: glucosylceramidase beta; IDE: insulin-degrading enzyme; LRRK2: leucine-rich repeat kinase 2; NFT: neurofibrillary tangle; PD: Parkinson’s disease; SNCA: alpha-synuclein; TFEB: transcription factor EB.
Autophagy in microglia of neurodegenerative diseases
AD: Microglia is a key contributor to amyloid-β (Aβ)-induced immune response in the propagation of neurodegeneration and synaptic dysfunction in AD (Guo et al., 2020). Various studies have recognized the interplay between microglial autophagy and neuroinflammation in AD. Autophagy in AD microglia negatively controls inflammasome activation and the generation of inflammatory cytokines. A study has shown that microglia-specific deletion of Atg7 shifts microglia towards a proinflammatory phenotype and enhances the production of pro-inflammatory cytokines (Xu et al., 2021). Houtman et al. (2019) showed an association of LC3-positive vesicles with NLRP3 aggregates in Becn1+/– microglia, indicating that autophagy modulates IL-1β and IL-18 production via NLRP3 activation in microglia.
Evidence suggests that autophagy in microglia regulates AD pathogenesis (Xu et al., 2021; Ou-Yang et al., 2023). Microglia mediates phagocytosis of a range of cellular debris, axonal fragments, Aβ and Tau deposits, and dying cells in AD (Gabandé-Rodríguez et al., 2020). Inference of microglial autophagy in synaptic function in young AD mouse hippocampus showed an anomalous accretion of autophagic vesicles associated with synaptic and axonal pathology (Sanchez-Varo et al., 2012). Studies have shown shared molecular pathways between phagocytosis and autophagy to establish a close crosstalk in microglia (Fu et al., 2014; Jülg et al., 2021). Microglia phagocytosis is coordinated by autophagy facilitators in several ways, such as LC3-associated phagocytosis (LAP) and LC3-associated endocytosis (LANDO) (Lee et al., 2019; Heckmann et al., 2019). During LAP, Atg5 and Atg7 mediate the LC3 recruitment to the single-membrane phagosomes (Martinez et al., 2011). Subsequently, LC3 and Beclin1 translocate to the phagosome membrane, enabling the fusion of lysosome with phagosome (Sanjuan et al., 2007). Such associations improve the efficiency of microglia phagocytosis in removing extracellular pathogens and debris (Plaza-Zabala et al., 2017). LANDO is a distinct but LAP-like process that helps in the clearance of Aβ and relieves neurodegeneration in murine models of AD (Heckmann et al., 2019). This study shows that the LANDO genes of Rubicon, Beclin1, Atg5, and Atg7 are crucial in recycling the Aβ receptors in microglia, e.g., triggering receptors expressed on myeloid cells 2 in the AD brain. Autophagy is also critical for the degradation of endocytosed aggregates in AD. In microglia-specific Atg7 knockout mice and Atg7 siRNA transfected microglia in vitro, impaired degradation of Aβ indicates the role of autophagy (Cho et al., 2014). Autophagy in microglia is also involved in Tau propagation as Atg7 deletion in microglia increases the intraneuronal Tau pathology (Xu et al., 2021). Furthermore, autophagy upstream regulatory mechanisms, such as energy homeostasis sensor AMPK, activate microglial autophagy in response to Aβ depositions and consequently lead to their lysosomal degradation (Cho et al., 2014). Injection of interferon-γ restores the autophagy activity in microglia by inhibiting the Akt/mTOR pathway in AD mice and promotes the Aβ clearance to rescue cognitive deficits (He et al., 2020). These studies show the protective functions of autophagy in microglia to slow AD progression through different mechanisms.
PD: PD is described by the progressive loss of dopaminergic neurons in the substantia nigra, accompanied by the accretion of alpha-synuclein (SNCA) in the form of Lewy bodies and Lewy neurites. Genome-wide association studies have identified the monogenetic causes of PD, including mutations in SNCA, leucine-rich repeat kinase 2 (LRRK2), Parkin RBR E3 ubiquitin protein ligase (PRKN), and PTEN induced kinase-1 (PINK1) (Chang et al., 2017; Nalls et al., 2019), which are all related to autophagy in normal brain. Interestingly, ablation of essential autophagy gene Atg5 in microglia induces PD-like symptoms in animals (Cheng et al., 2020). Additionally, depleting microglial autophagy in a PD mouse model accelerates the loss of dopaminergic neurons, implying the influence of microglial autophagy in PD pathogenesis (Choi et al., 2022). These studies indicate that autophagy in microglia is involved in the initiation and progression of PD. Mechanistically, Atg5 deletion exacerbates the pro-inflammatory responses and aggravates the MPTP-induced neurodegeneration by activating NLRP3 inflammasome (Qin et al., 2021). Studies have shown that an excessive stimulation of NLRP3 inflammasome might worsen autophagy in microglia and further exacerbates the pathogenesis of neurodegenerative diseases. Atg7-deletion in microglia reveals an enhanced level of insoluble and phosphorylated SNCA (p-S129) in the PD brain (Choi et al., 2020). Selective autophagy, facilitated by autophagy receptor p62/SQSTM1 in microglia suppresses the accumulation of misfolded/aggregated SNCA (termed synucleinphagy), suggesting a protective role of microglia autophagy in the clearing of SNCA (Choi et al., 2020). Aggregations of SNCA trigger the activation of NLRP3 inflammasome in microglia through collaboration with Toll-like receptors and activation of nuclear factor-κB (Panicker et al., 2019). For the initiation of synucleinphagy, microglia Toll-like receptor 4 stimulates the transcriptional upregulation of p62/SQSTM1 through the nuclear factor-κB signaling pathway that further promotes the degradation of SNCA (Choi et al., 2020). Moreover, SNCA is also a substrate of CMA. Thus, enhancing CMA could reduce the SNCA levels and protect cells from SNCA-induced neurotoxicity (Xilouri et al., 2013). Chen et al. (2021) have explored the activation of microglial autophagy stimulated by the p38-transcription factor EB (TFEB) pathway by inhibiting CMA-dependent NLRP3 degradation, which might be used as a therapeutic strategy for PD patients. Converging evidence has shown that microglial autophagy is neuroprotective in PD.
Autophagy in astrocytes of neurodegenerative diseases
AD: Astrocytes promote the diminution of extracellular Aβ and prevent its toxicity by supporting its internalization and degradation in astrocytes (Phatnani and Maniatis, 2015). In astrocyte culture exposed to Aβ1–42 peptide, Aβ content is mainly observed in LC3-labelled autophagosomes. Atg5 deletion interrupts the autophagic flux, and LC3 overlapping with Aβ accretion shows a significant reduction (Pomilio et al., 2016). ApoE promotes the astrocytes ability for Aβ clearing process in the AD brain (Husain et al., 2021). It is suggested that autophagy is essential for ApoE-dependent Aβ clearance in astrocytes (Simonovitch et al., 2016). In response to Aβ, mouse primary astrocytes release insulin-degrading enzyme through an autophagy-dependent process. Insulin-degrading enzyme is an essential protease involved in the degradation of Aβ in the extracellular space. In Atg7 gene ablated mice, the level of insulin-degrading enzyme is reduced, suggesting the involvement of astrocyte autophagy in the degradation of extracellular Aβ (Son et al., 2016). TFEB is a well-known transcriptional factor that organizes the autophagosome-lysosome function by increasing the expression of genes involved in lysosome biogenesis, autophagosome formation, and lysosome function (Cortes et al., 2019). Intriguingly, virus-aided astrocyte-selective overexpression of TFEB augments the lysosomal function and reduces Aβ plaque deposition in the hippocampus of transgenic mice with mutant human APP and PSEN1 genes (Xiao et al., 2014). In astrocyte culture, expression of exogenous TFEB increases the uptake, trafficking, and degradation of Aβ, suggesting an autolysosome/endolysosomal pathway begun by astrocytes for Aβ clearance in AD (Xiao et al., 2014). These observations for TFEB in astrocytes provide a link between autophagy and Aβ clearance.
PD: The neuropathology of PD includes dysfunctional protein degradation and SNCA, neuroinflammation, glia activation, oxidative stress, and mitochondrial dysfunction. Though PD has been regarded as a dopaminergic neuron disease, the astrocyte’s contribution to PD pathogenesis is gaining attention. Reactive astrocytes with enhanced expression of SNCA and dysfunctional mitochondria in astrocytes from PD patients show disease pathology (Bantle et al., 2019; Wang et al., 2021). Dysfunctional proteolysis is also considered one of the contributing factors in PD. In astrocytes, the autophagy-lysosomal pathway and ubiquitin-proteasome system begin clearing and refolding of abnormal proteins. Alterations in autophagic pathways might halt the accretion of protease-resistant misfolded and aggregated proteins and damaged organelles in PD (Galves et al., 2019). It has been shown that lysosomal inhibitor bafilomycin A1 improves the formation of detergent-insoluble SNCA in astrocytes (Lee et al., 2010). A similar effect is also observed with autophagy inhibitor 3-methyladenine, as it reverses the recovery of the degradative ability of astrocytes and reduces the level of intracellular SNCA engulfed by astrocytes (Hua et al., 2019). A selective inhibition of autophagy in astrocytes through overexpression of αβ-crystallin increases the SNCA accumulated in the brains of transgenic mice expressing the human SNCA A30P mutant (Lu et al., 2019). Recently, there are a number of genes linked to autophagy and astrocytes as risk factors in PD, such as glucocerebrosidase, LRRK2, PINK1, ATP13A2, PARK2, and PARK7 (Booth et al., 2017). A study has shown that LRRK2 G2019S mutation in PD patient’s iPSC-derived astrocytes caused altered CMA and impaired macroautophagy that further leads to advanced endogenous SNCA accretion (di Domenico et al., 2019). Furthermore, ablation of astrocyte mitophagy by knockout Kir6.1 causes the enhanced degeneration of dopaminergic neurons in substantia nigra pars compacta and leads to acute motor dysfunctions in MPTP stimulated PD model. Restoring the mitophagy in astrocytes supports dopaminergic neuron survival and mitochondrial functions, suggesting a crucial role for astrocyte mitophagy in PD pathogenesis (Hu et al., 2019). Astrocytes deficient in Pink1 and Parkin showed mitophagy defects that caused a reduced distribution of astrocytes in the substantia nigra. Lack of astrocyte-mediated restoration contributed to the progression of PD (Kano et al., 2020). Another PD-allied gene, glucocerebrosidase mutation in astrocytes leads to dysfunctional proteolysis by affecting the autophagy-lysosome pathway that creates excessive neuronal SNCA accumulation in PD (Aflaki et al., 2020).
Autophagy in oligodendrocytes of neurodegenerative injuries
Numerous previous studies have documented that autophagy in oligodendrocytes is involved in both brain injury and recovery (Belgrad et al., 2020). Conditional ablation of Atg5 in oligodendrocytes reduces recovery after spinal cord injury (Ohri et al., 2018). In traumatic brain injury, stimulation of autophagy reduces the extent of the damage for grey and white matter in both the cortex and hippocampus (Yin et al., 2018). Autophagy not only plays an empirical role in the differentiation and survival of oligodendrocytes (Bankston et al., 2019) but also impacts its myelination ability. In multiple sclerosis, alterations in oligodendrocyte autophagy affect myelin plasticity, leading to cytoplasm decompaction and reduced myelin wraps, which eventually trigger demyelination (Belgrad et al., 2020). Moreover, a recent study has shown that deactivation of oligodendroglia autophagy by Atg7 deletion causes adult-onset increases in myelin, enhanced myelin sheath width, and irregular myelin structures that ultimately impair neural function, concluding in adult-onset advanced motor decline, neurodegeneration, and death (Aber et al., 2022). Better understanding of the underlying autophagy mechanisms in oligodendrocytes demands pursual as it will provide the direction for developing the treatments against demyelinated brain injuries.
Therapeutic Implication of Autophagy Modulation in Glia
Growing evidence shows that autophagy dysregulation in glia contributes to various neurodegenerative diseases. Despite these studies, very few approaches have been made to target the autophagy pathways in glia as therapeutic strategies for treatment. In a transgenic mouse model for AD-related amyloidosis (CRND8), enhancement of the autophagy/lysosomal pathway through single-walled carbon nanotubes aids in clearing amyloid (Xue et al., 2014). A study conducted on a human glia cell line reported that an increase in BAG3-dependent autophagy by mitigating the heat shock protein CRYAB increased the removal of α-synuclein (Lu et al., 2019). More specifically, modulation of autophagic genes in astrocytes, like adeno-associated virus (AAV) mediated TFEB to increase its expression in neurons, helping to reduce the Aβ levels and amyloid accretion in a mouse model expressing mutant human PSEN1 and APP genes (Xiao et al., 2014). The AAV-mediated cell-specific gene delivery was also used for TFEB expression in astrocytes, which shows promising effects to increase uptake of extracellular tau species and reduce tau spreading in the PS19 mouse model (Martini-Stoica et al., 2018). The efficiency of AAV delivery in microglia used to be a bottleneck for in vivo experiments. Recent progress (Lin et al., 2022; Okada et al., 2022) for specific and efficient delivery of autophagy genes or regulator genes such as TFEB may overcome this barrier for future therapy in AD and PD. Overall, it is a promising strategy to develop specific and efficient AAV gene therapies to deliver autophagy-related genes to glia in neurodegenerative and neurodevelopmental disorders.
Conclusions and Perspectives
Autophagy is evolving as a critical process that impacts the human brain throughout the lifespan in both physiological and pathological conditions. Ample results from recent studies have discussed the impairments of autophagy in various disorders in developing, adult, and aging brains. However, these studies are mainly attributed to neuronal autophagy. In recent years, increasing evidence is available indicating glial autophagy dysfunction in various brain pathologies. Involvement of autophagy in the regulation of glial functions is an intriguing area of research as it not only affects glial health by modulating the dead/damaged cell debris clearance but also contributes to signaling mediators through proteolytic processing. Thus, the efficiency of glial autophagy might have profound implications on health, function, and response to various stress stimuli in the brain. Nevertheless, the information on the elaborate interaction between glia and autophagy genes in brain homeostasis and brain disorders is still far from being done. The investigation of glial autophagy regulation is indispensable from both scientific and clinical standpoints. Exploration of the molecular mechanisms of autophagy pathways in glia is vital to a comprehensive platform for neurotherapeutic approaches. Similarly, during brain development autophagy plays a crucial role in genesis and differentiation of the NSCs. Human clinical and genetic studies have reported the contribution of autophagy-related genes in congenital mutations in various neurodevelopmental disorders. Autophagy regulates multitasking attributes of NSCs, and this aspect also deserves a more vigorous investigative approach to develop a better insight into the contribution of specific autophagy genes in the maintenance and regulation of NSCs as well as their cell fates in developing, adult, and aging brains.
Additional file: Open peer review report 1 (78.8KB, pdf) .
Funding Statement
Funding: The work is supported by NIH R01NS103981 and R01CA273586 (to CW).
Footnotes
Open peer reviewer: Qian Cai, Rutgers, The State University of New Jersey, USA.
P-Reviewer: Cai Q; C-Editors: Zhao M, Sun Y, Qiu Y; T-Editor: Jia Y
Conflicts of interest: The authors declare no conflicts of interest.
Data availability statement: The data are available from the corresponding author on reasonable request.
References
- 1.Aber ER, Griffey CJ, Davies T, Li AM, Yang YJ, Croce KR, Goldman JE, Grutzendler J, Canman JC, Yamamoto A. Oligodendroglial macroautophagy is essential for myelin sheath turnover to prevent neurodegeneration and death. Cell Rep. 2022;41:111480. doi: 10.1016/j.celrep.2022.111480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aflaki E, Stubblefield BK, McGlinchey RP, McMahon B, Ory DS, Sidransky E. A characterization of Gaucher iPS-derived astrocytes: potential implications for Parkinson's disease. Neurobiol Dis. 2020;134:104647. doi: 10.1016/j.nbd.2019.104647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alam MM, Zhao XF, Liao Y, Mathur R, McCallum SE, Mazurkiewicz JE, Adamo MA, Feustel P, Belin S, Poitelon Y, Zhu XC, Huang Y. Deficiency of microglial autophagy increases the density of oligodendrocytes and susceptibility to severe forms of seizures. eNeuro 8. 2021 doi: 10.1523/ENEURO.0183-20.2021. ENEURO.0183-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Beltagi M. Autism medical comorbidities. World J Clin Pediatr. 2021;10:15–28. doi: 10.5409/wjcp.v10.i3.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bankston AN, Forston MD, Howard RM, Andres KR, Smith AE, Ohri SS, Bates ML, Bunge MB, Whittemore SR. Autophagy is essential for oligodendrocyte differentiation, survival and proper myelination. Glia. 2019;67:1745–1759. doi: 10.1002/glia.23646. [DOI] [PubMed] [Google Scholar]
- 6.Bantle CM, Hirst WD, Weihofen A, Shlevkov E. Mitochondrial dysfunction in astrocytes: a role in Parkinson's disease? Front Cell Dev Biol. 2021;8:608026. doi: 10.3389/fcell.2020.608026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belgrad J, De Pace R, Fields RD. Autophagy in myelinating glia. J Neurosci. 2020;40:256–266. doi: 10.1523/JNEUROSCI.1066-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund R, Guerreiro-Cacais AO, Adzemovic MZ, Zeitelhofer M, Lund H, Ewing E, Ruhrmann S, Nutma E, Parsa R, Thessen-Hedreul M, Amor S, Harris RA, Olsson T, Jagodic M. Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci Immunol. 2020;5:eabb5077. doi: 10.1126/sciimmunol.abb5077. [DOI] [PubMed] [Google Scholar]
- 9.Booth HDE, Hirst WD, Wade-Martins R. The role of astrocyte dysfunction in Parkinson's disease pathogenesis. Trends Neurosci. 2017;40:358–370. doi: 10.1016/j.tins.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borst K, Dumas AA, Prinz M. Microglia: immune and non-immune functions. Immunity. 2021;54:2194–2208. doi: 10.1016/j.immuni.2021.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Butler CA, Popescu AS, Kitchener EJA, Allendorf DH, Puigdellívol M, Brown GC. Microglial phagocytosis of neurons in neurodegeneration, and its regulation. J Neurochem. 2021;158:621–639. doi: 10.1111/jnc.15327. [DOI] [PubMed] [Google Scholar]
- 12.Byrne S, Jansen L, U-King-Im J-M, Siddiqui A, Lidov H, Bodi I. EPG5-related Vici syndrome: a paradigm of neurodevelopmental disorders with defective autophagy. Brain. 2016;139:765–781. doi: 10.1093/brain/awv393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrasekaran A, Dittlau KS, Corsi GI, Haukedal H, Doncheva NT, Ramakrishna S, Ambardar S, Salcedo C, Schmidt SI, Zhang Y, Cirera S, Pihl M, Schmid B, Nielsen TT, Nielsen JE, Kolko M, Kobolák J, Dinnyés A, Hyttel P, Palakodeti D, et al. Astrocytic reactivity triggered by defective autophagy and metabolic failure causes neurotoxicity in frontotemporal dementia type 3. Stem Cell Reports. 2021;16:2736–2751. doi: 10.1016/j.stemcr.2021.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Mao K, Yu H, Wen Y, She H, Zhang H, Liu L, Li M, Li W, Zou F. p38-TFEB pathways promote microglia activation through inhibiting CMA-mediated NLRP3 degradation in Parkinson's disease. J Neuroinflammation. 2021;18:295. doi: 10.1186/s12974-021-02349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, He Y, Lu F. Autophagy in stem cell biology: a perspective on stem cell self-renewal and differentiation. Stem Cells Int. 2018;2018:9131397. doi: 10.1155/2018/9131397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng J, Liao Y, Dong Y, Hu H, Yang N, Kong X, Li S, Li X, Guo J, Qin L, Yu J, Ma C, Li J, Li M, Tang B, Yuan Z. Microglial autophagy defect causes Parkinson disease-like symptoms by accelerating inflammasome activation in mice. Autophagy. 2020;16:2193–2205. doi: 10.1080/15548627.2020.1719723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho KS, Lee JH, Cho J, Cha GH, Song GJ. Autophagy modulators and neuroinflammation. Curr Med Chem. 2020;27:955–982. doi: 10.2174/0929867325666181031144605. [DOI] [PubMed] [Google Scholar]
- 18.Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, Yoon SY. Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy. 2014;10:1761–1775. doi: 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi I, Zhang Y, Seegobin SP, Pruvost M, Wang Q, Purtell K, Zhang B, Yue Z. Microglia clear neuron-released α-synuclein via selective autophagy and prevent neurodegeneration. Nat Commun. 2020;11:1386. doi: 10.1038/s41467-020-15119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi I, Heaton GR, Lee YK, Yue Z. Regulation of α-synuclein homeostasis and inflammasome activation by microglial autophagy. Sci Adv. 2022;8:eabn1298. doi: 10.1126/sciadv.abn1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collier JJ, Guissart C, Oláhová M, Sasorith S, Piron-Prunier F, Suomi F, Zhang D, Martinez-Lopez N, Leboucq N, Bahr A, Azzarello-Burri S, Reich S, Schöls L, Polvikoski TM, Meyer P, Larrieu L, Schaefer AM, Alsaif HS, Alyamani S, Zuchner S, et al. Developmental consequences of defective ATG7-mediated autophagy in humans. N Engl J Med. 2021;384:2406–2417. doi: 10.1056/NEJMoa1915722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortes CJ, La Spada AR. TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: molecular mechanisms, cellular processes, and emerging therapeutic opportunities. Neurobiol Dis. 2019;122:83–93. doi: 10.1016/j.nbd.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng Z, Zhou X, Lu JH, Yue Z. Autophagy deficiency in neurodevelopmental disorders. Cell Biosci. 2021;11:214. doi: 10.1186/s13578-021-00726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.di Domenico A, Carola G, Calatayud C, Pons-Espinal M, Muñoz JP, Richaud-Patin Y, Fernandez-Carasa I, Gut M, Faella A, Parameswaran J, Soriano J, Ferrer I, Tolosa E, Zorzano A, Cuervo AM, Raya A, Consiglio A. Patient-specific iPSC-derived astrocytes contribute to non-cell-autonomous neurodegeneration in Parkinson's disease. Stem Cell Reports. 2019;12:213–229. doi: 10.1016/j.stemcr.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diot A, Morten K, Poulton J. Mitophagy plays a central role in mitochondrial ageing. Mamm Genome. 2016;27:381–395. doi: 10.1007/s00335-016-9651-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doetsch F. A niche for adult neural stem cells. Curr Opin Genet Dev. 2003;13:543–550. doi: 10.1016/j.gde.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon MS, Eenjes E, Bosco JR, Fox LM, Lystad AH, Oo TF, Yarygina O, Mita T, Waguri S, Ichimura Y, Komatsu M, Simonsen A, Burke RE, Mason CA, Yamamoto A. Autophagy linked FYVE (Alfy/ WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife. 2016;5:e14810. doi: 10.7554/eLife.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 31.Fernández ÁF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, Marciano DK, Schiattarella GG, Bhagat G, Moe OW, Hu MC, Levine B. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filippelli RL, Kamyabiazar S, Chang NC. Monitoring autophagy in neural stem and progenitor cells. Methods Mol Biol. 2022;2515:99–116. doi: 10.1007/978-1-0716-2409-8_7. [DOI] [PubMed] [Google Scholar]
- 33.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 34.Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S. Inflammaging and 'Garb-aging'. Trends Endocrinol Metab. 2017;28:199–212. doi: 10.1016/j.tem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Gabandé-Rodríguez E, Keane L, Capasso M. Microglial phagocytosis in aging and Alzheimer's disease. J Neurosci Res. 2020;98:284–298. doi: 10.1002/jnr.24419. [DOI] [PubMed] [Google Scholar]
- 36.Ganley IG, Lam dH, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galves M, Rathi R, Prag G, Ashkenazi A. Ubiquitin signaling and degradation of aggregate-prone proteins. Trends Biochem Sci. 2019;44:872–884. doi: 10.1016/j.tibs.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Germic N, Frangez Z, Yousefi S, Simon HU. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ. 2019;26:715–727. doi: 10.1038/s41418-019-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González A, Hall MN, Lin SC, Hardie DG. AMPK and TOR: The Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 2020;31:472–492. doi: 10.1016/j.cmet.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Guo T, Zhang D, Zeng Y, Huang TY, Xu H, Zhao Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer's disease. Mol Neurodegener. 2020;15:40. doi: 10.1186/s13024-020-00391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha S, Jeong SH, Yi K, Chu JJM, Kim S, Kim EK, Yu SW. Autophagy mediates astrogenesis in adult hippocampal neural stem cells. Exp Neurobiol. 2019;28:229–246. doi: 10.5607/en.2019.28.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Z, Yang Y, Xing Z, Zuo Z, Wang R, Gu H, Qi F, Yao Z. Intraperitoneal injection of IFN-γrestores microglial autophagy, promotes amyloid-βclearance and improves cognition in APP/PS1 mice. Cell Death Dis. 2020;11:440. doi: 10.1038/s41419-020-2644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell. 2019;178:536–551. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henn RE, Noureldein MH, Elzinga SE, Kim B, Savelieff MG, Feldman EL. Glial-neuron crosstalk in health and disease: A focus on metabolism, obesity and cognitive impairment. Neurobiol Dis. 2022;170:105766. doi: 10.1016/j.nbd.2022.105766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollenstein DM, Kraft C. Autophagosomes are formed at a distinct cellular structure. Curr Opin Cell Biol. 2020;65:50–57. doi: 10.1016/j.ceb.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houtman J, Freitag K, Gimber N, Schmoranzer J, Heppner FL, Jendrach M. Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J. 2019;38:e99430. doi: 10.15252/embj.201899430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu ZL, Sun T, Lu M, Ding JH, Du RH, Hu G. Kir6.1/K-ATP channel on astrocytes protects against dopaminergic neurodegeneration in the MPTP mouse model of Parkinson's disease via promoting mitophagy. Brain Behav Immun. 2019;81:509–522. doi: 10.1016/j.bbi.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Hua J, Yin N, Xu S, Chen Q, Tao T, Zhang J, Ding J, Fan Y, Hu G. Enhancing the astrocytic clearance of extracellular α-synuclein aggregates by ginkgolides attenuates neural cell injury. Cell Mol Neurobiol. 2019;39:1017–1028. doi: 10.1007/s10571-019-00696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hui KK, Takashima N, Watanabe A, Chater TE, Matsukawa H, Nekooki-Machida Y, Nilsson P, Endo R, Goda Y, Saido TC, Yoshikawa T, Tanaka M. GABARAPs dysfunction by autophagy deficiency in adolescent brain impairs GABA(A) receptor trafficking and social behavior. Sci Adv. 2019;5:eaau8237. doi: 10.1126/sciadv.aau8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Husain MA, Laurent B, Plourde M. APOE and Alzheimer's disease: from lipid transport to physiopathology and therapeutics. Front Neurosci. 2021;15:630502. doi: 10.3389/fnins.2021.630502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jänen SB, Chaachouay H, Richter-Landsberg C. Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia. 2010;58:1766–1774. doi: 10.1002/glia.21047. [DOI] [PubMed] [Google Scholar]
- 53.Jean F, Stuart A, Tarailo-Graovac M. Dissecting the genetic and etiological causes of primary microcephaly. Front Neurol. 2020;11:570830. doi: 10.3389/fneur.2020.570830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang GM, Tan Y, Wang H, Peng L, Chen HT, Meng XJ, Li LL, Liu Y, Li WF, Shan H. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol Cancer. 2019;18:17. doi: 10.1186/s12943-019-0944-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin MM, Wang F, Qi D, Liu WW, Gu C, Mao CJ, Yang YP, Zhao Z, Hu LF, Liu CF. A critical role of autophagy in regulating microglia polarization in neurodegeneration. Front Aging Neurosci. 2018;10:378. doi: 10.3389/fnagi.2018.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jülg J, Strohm L, Behrends C. Canonical and noncanonical autophagy pathways in microglia. Mol Cell Biol. 2021;41:e0038920. doi: 10.1128/MCB.00389-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung S, Choe S, Woo H, Jeong H, An HK, Moon H, Ryu HY, Yeo BK, Lee YW, Choi H, Mun JY, Sun W, Choe HK, Kim EK, Yu SW. Autophagic death of neural stem cells mediates chronic stress-induced decline of adult hippocampal neurogenesis and cognitive deficits. Autophagy. 2020b;16:512–530. doi: 10.1080/15548627.2019.1630222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung S, Jeong H, Yu SW. Autophagy as a decisive process for cell death. Exp Mol Med. 2020a;52:921–930. doi: 10.1038/s12276-020-0455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadir R, Harel T, Markus B, Perez Y, Bakhrat A, Cohen I, Volodarsky M, Feintsein-Linial M, Chervinski E, Zlotogora J, Sivan S, Birnbaum RY, Abdu U, Shalev S, Birk OS. ALFY-controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genet. 2016;12:e1005919. doi: 10.1371/journal.pgen.1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kano M, Takanashi M. Reduced astrocytic reactivity in human brains and midbrain organoids with PRKN mutations. NPJ Parkinsons Dis. 2020;6:33. doi: 10.1038/s41531-020-00137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawabata T, Yoshimori T. Autophagosome biogenesis and human health. Cell Discov. 2020;6:33. doi: 10.1038/s41421-020-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim HJ, Cho MH, Shim WH, Kim JK, Jeon EY, Kim DH, Yoon SY. Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol Psychiatry. 2017;22:1576–1584. doi: 10.1038/mp.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Schulman BA, Xu J, Semple I, Ro SH, Kim B, Mavioglu RN, Tolun A, Jipa A, Takats S, Karpati M, Li JZ, Yapici Z, Juhasz G, Lee JH, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife. 2016;5:e12245. doi: 10.7554/eLife.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le Duc D, Giulivi C, Hiatt SM, Napoli E, Panoutsopoulos A, Harlan De Crescenzo A, Kotzaeridou U, Syrbe S, Anagnostou E, Azage M, Bend R, Begtrup A, Brown NJ, Büttner B, Cho MT, Cooper GM, Doering JH, Dubourg C, Everman DB, Hildebrand MS, et al. Pathogenic WDFY3 variants cause neurodevelopmental disorders and opposing effects on brain size. Brain. 2019;142:2617–2630. doi: 10.1093/brain/awz198. Erratum in: Brain 142: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee HJ, Suk JE, Patrick C, Bae EJ, Cho JH, Rho S, Hwang D, Masliah E, Lee SJ. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285:9262–9272. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JW, Nam H, Kim LE, Jeon Y, Min H, Ha S, Lee Y, Kim SY, Lee SJ, Kim EK, Yu SW. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy. 2019;15:753–770. doi: 10.1080/15548627.2018.1556946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G, Sherchan P, Tang Z, Tang J. Autophagy &phagocytosis in neurological disorders and their possible cross-talk. Curr Neuropharmacol. 2021;19:1912–1924. doi: 10.2174/1570159X19666210407150632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li M, Lu G, Hu J, Shen X, Ju J, Gao Y, Qu L, Xia Y, Chen Y, Bai Y. EVA1A/TMEM166 regulates embryonic neurogenesis by autophagy. Stem Cell Reports. 2016;6:396–410. doi: 10.1016/j.stemcr.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin CH, Wu YR, Yang JM, Chen WL, Chao CY, Chen IC, Lin TH, Wu YC, Hsu KC, Chen CM, Lee GC, Hsieh-Li HM, Lee CM, Lee-Chen GJ. Novel lactulose and melibiose targeting autophagy to reduce polyQ aggregation in cell models of spinocerebellar ataxia 3. CNS Neurol Disord Drug Targets. 2016;15:351–359. doi: 10.2174/1871527314666150821101522. [DOI] [PubMed] [Google Scholar]
- 74.Lin R, Zhou Y, Yan T, Wang R, Li H, Wu Z, Zhang X, Zhou X, Zhao F, Zhang L, Li Y, Luo M. Directed evolution of adeno-associated virus for efficient gene delivery to microglia. Nat Methods. 2022;19:976–985. doi: 10.1038/s41592-022-01547-7. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Wang C, Yi F, Yeo S, Haas M, Tang X, Guan JL. Non-canonical function of FIP200 is required for neural stem cell maintenance and differentiation by limiting TBK1 activation and p62 aggregate formation. Sci Rep. 2021;11:23907. doi: 10.1038/s41598-021-03404-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Llorente V, Velarde P, Desco M, Gómez-Gaviro MV. Current understanding of the neural stem cell niches. Cells. 2022;11:3002. doi: 10.3390/cells11193002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu G, Wang Y, Shi Y, Zhang Z, Huang C, He W, Wang C, Shen HM. Autophagy in health and disease: From molecular mechanisms to therapeutic target. MedComm (2020) 2022;3:e150. doi: 10.1002/mco2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu SZ, Guo YS, Liang PZ, Zhang SZ, Yin S, Yin YQ, Wang XM, Ding F, Gu XS, Zhou JW. Suppression of astrocytic autophagy by αB-crystallin contributes to α-synuclein inclusion formation. Transl Neurodegener. 2019;8:3. doi: 10.1186/s40035-018-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martini-Stoica H, Cole AL, Swartzlander DB, Chen F, Wan YW, Bajaj L, Bader DA, Lee VMY, Trojanowski JQ, Liu Z, Sardiello M, Zheng H. TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J Exp Med. 2018;215:2355–2377. doi: 10.1084/jem.20172158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Metur SP, Klionsky DJ. Adaptive immunity at the crossroads of autophagy and metabolism. Cell Mol Immunol. 2021;18:1096–1105. doi: 10.1038/s41423-021-00662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minchev D, Kazakova M, Sarafian V. Neuroinflammation and autophagy in Parkinson's disease—novel perspectives. Int J Mol Sci. 2022;23:14997. doi: 10.3390/ijms232314997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Motori E, Puyal J, Toni N, Ghanem A, Angeloni C, Malaguti M, Cantelli-Forti G, Berninger B, Conzelmann KK, Götz M, Winklhofer KF, Hrelia S, Bergami M. Inflammation-induced alteration of astrocyte mitochondrial dynamics requires autophagy for mitochondrial network maintenance. Cell Metab. 2013;18:844–859. doi: 10.1016/j.cmet.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 86.Münz C. Canonical and non-canonical functions of the autophagy machinery in MHC restricted antigen presentation. Front Immunol. 2022;13:868888. doi: 10.3389/fimmu.2022.868888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, von Coelln R, Simón-Sánchez J, Schulte C, Sharma M, Krohn L, Pihlstrøm L, Siitonen A, Iwaki H, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Obernier K, Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146:dev156059. doi: 10.1242/dev.156059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohri S, Bankston AN, Mullins SA, Liu Y, Andres KR, Beare JE, oward RM, Burke DA, Riegler AS, Smith AE, Hetman M, Whittemore SR. Blocking autophagy in oligodendrocytes limits functional recovery after spinal cord injury. J Neurosci. 2018;38:5900–5912. doi: 10.1523/JNEUROSCI.0679-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okada Y, Hosoi N, Matsuzaki Y, Fukai Y, Hiraga A, Nakai J, Nitta K, Shinohara Y, Konno A, Hirai H. Development of microglia-targeting adeno-associated viral vectors as tools to study microglial behavior in vivo. Commun Biol. 2022;5:1224. doi: 10.1038/s42003-022-04200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ordureau A, Kraus F, Zhang J, An H, Park S, Ahfeldt T, Paulo JA, Harper JW. Temporal proteomics during neurogenesis reveals large-scale proteome and organelle remodeling via selective autophagy. Mol Cell. 2021;81:5082–5098. doi: 10.1016/j.molcel.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ou-Yang P, Cai ZY, Zhang ZH. Molecular regulation mechanism of microglial autophagy in the pathology of Alzheimer's disease. Aging Dis. 2023 doi: 10.14336/AD.2023.0106. doi: 10.14336/AD.2023.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, Hiller D, Jiang S, Protopopov A, Wong WH, Chin L, Ligon KL, DePinho RA. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Panicker N, Sarkar S, Harischandra DS, Neal M, Kam TI, Jin H, Saminathan H, Langley M, Charli A, Samidurai M, Rokad D, Ghaisas S, Pletnikova O, Dawson VL, Dawson TM, Anantharam V, Kanthasamy AG, Kanthasamy A. Fyn kinase regulates misfolded α-synuclein uptake and NLRP3 inflammasome activation in microglia. J Exp Med. 2019;216:1411–1430. doi: 10.1084/jem.20182191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pavone P, Praticò AD, Pavone V, Lubrano R, Falsaperla R, Rizzo R, Ruggieri M. Ataxia in children: early recognition and clinical evaluation. Ital J Pediatr. 2017;43:6. doi: 10.1186/s13052-016-0325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Phatnani H, Maniatis T. Astrocytes in neurodegenerative disease. Cold Spring Harb Perspect Biol. 2015;7:a020628. doi: 10.1101/cshperspect.a020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Plaza-Zabala A, Sierra-Torre V, Sierra A. Autophagy and microglia: novel partners in neurodegeneration and aging. Int J Mol Sci. 2017;18:598. doi: 10.3390/ijms18030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pomilio C, Pavia P, Gorojod RM, Vinuesa A, Alaimo A, Galvan V, Kotler ML, Beauquis J, Saravia F. Glial alterations from early to late stages in a model of Alzheimer's disease: Evidence of autophagy involvement in Aβinternalization. Hippocampus. 2016;26:194–210. doi: 10.1002/hipo.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y, De Rubeis S, Durand S, Stevens C, Rehnstrom K, Palotie A, Daly MJ, Ma'ayan A, Fromer M, Buxbaum JD. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet. 2013;93:607–619. doi: 10.1016/j.ajhg.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pradel B, Robert-Hebmann V, Espert L. Regulation of innate immune responses by autophagy: a goldmine for viruses. Front Immunol. 2020;11:578038. doi: 10.3389/fimmu.2020.578038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qin Y, Qiu J, Wang P, Liu J, Zhao Y, Jiang F, Lou H. Impaired autophagy in microglia aggravates dopaminergic neurodegeneration by regulating NLRP3 inflammasome activation in experimental models of Parkinson's disease. Brain Behav Immun. 2021;91:324–338. doi: 10.1016/j.bbi.2020.10.010. [DOI] [PubMed] [Google Scholar]
- 102.Rodríguez-Bodero A, Encinas-Pérez JM. Does the plasticity of neural stem cells and neurogenesis make them biosensors of disease and damage? Front Neurosci. 2022;16:977209. doi: 10.3389/fnins.2022.977209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sakai S, Shichita T. Inflammation and neural repair after ischemic brain injury. Neurochem Int. 2019;130:104316. doi: 10.1016/j.neuint.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 105.Sanchez-Varo R, Trujillo-Estrada L, Sanchez-Mejias E, Torres M, Baglietto-Vargas D, Moreno-Gonzalez I, De Castro V, Jimenez S, Ruano D, Vizuete M, Davila JC, Garcia-Verdugo JM, Jimenez AJ, Vitorica J, Gutierrez A. Abnormal accumulation of autophagic vesicles correlates with axonal and synaptic pathology in young Alzheimer's mice hippocampus. Acta Neuropathol. 2012;123:53–70. doi: 10.1007/s00401-011-0896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The heritability of autism spectrum disorder. JAMA. 2017;318:1182–1184. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 108.Sarret C. Leukodystrophies and genetic leukoencephalopathies in children. Revue Neurologique. 2020;176:10–19. doi: 10.1016/j.neurol.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 109.Sato A, Kasai S, Kobayashi T, Takamatsu Y, Hino O, Ikeda K, Mizuguchi M. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat Commun. 2012;3:1292. doi: 10.1038/ncomms2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schäffner I, Minakaki G, Khan MA, Balta EA, Schlötzer-Schrehardt U, Schwarz TJ, Beckervordersandforth R, Winner B, Webb AE, DePinho RA, Paik J, Wurst W, Klucken J, Lie DC. FoxO function is essential for maintenance of autophagic flux and neuronal morphogenesis in adult neurogenesis. Neuron. 2018;99:1188–1203. doi: 10.1016/j.neuron.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Simonovitch S, Schmukler E, Bespalko A, Iram T, Frenkel D, Holtzman DM, Masliah E, Michaelson DM, Pinkas-Kramarski R. Impaired autophagy in APOE4 astrocytes. J Alzheimers Dis. 2016;51:915–927. doi: 10.3233/JAD-151101. [DOI] [PubMed] [Google Scholar]
- 113.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sofroniew MV. Astrocyte reactivity: subtypes, states and functions in CNS innate immunity. Trends Immunol. 2020;41:758–770. doi: 10.1016/j.it.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Son SM, Cha MY, Choi H, Kang S, Choi H, Lee MS, Park SA, Mook-Jung I. Insulin-degrading enzyme secretion from astrocytes is mediated by an autophagy-based unconventional secretory pathway in Alzheimer disease. Autophagy. 2016;12:784–800. doi: 10.1080/15548627.2016.1159375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stranks AJ, Hansen AL, Panse I, Mortensen M, Ferguson DJ, Puleston DJ, Shenderov K, Watson AS, Veldhoen M, Phadwal K, Cerundolo V, Simon AK. Autophagy controls acquisition of aging features in macrophages. J Innate Immun. 2015;7:375–391. doi: 10.1159/000370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun J, Song Y, Chen Z, Qiu J, Zhu S, Wu L, Xing L. Heterogeneity and molecular markers for CNS glial cells revealed by single-cell transcriptomics. Cell Mol Neurobiol. 2022;42:2629–2642. doi: 10.1007/s10571-021-01159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takahama M, Akira S, Saitoh T. Autophagy limits activation of the inflammasomes. Immunol Rev. 2018;281:62–73. doi: 10.1111/imr.12613. [DOI] [PubMed] [Google Scholar]
- 121.Tang G, Yue Z, Talloczy Z, Hagemann T, Cho W, Messing A, Sulzer DI, Goldman JE. Autophagy induced by Alexander disease-mutant GFAP accumulation is regulated by p38/MAPK and mTOR signaling pathways. Hum Mol Genet. 2008;17:1540–1555. doi: 10.1093/hmg/ddn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thellung S, Scoti B, Corsaro A, Villa V, Nizzari M, Gagliani MC, Porcile C, Russo C, Pagano A, Tacchetti C, Cortese K, Florio T. Pharmacological activation of autophagy favors the clearing of intracellular aggregates of misfolded prion protein peptide to prevent neuronal death. Cell Death Dis. 2018;9:166. doi: 10.1038/s41419-017-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vainchtein ID, Molofsky AV. Astrocytes and microglia: in sickness and in health. Trends Neurosci. 2020;43:144–154. doi: 10.1016/j.tins.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vanderver A. Genetic leukoencephalopathies in adults. Continuum (Minneap Minn) 2016;22:916–942. doi: 10.1212/CON.0000000000000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vázquez P, Arroba A, I, Cecconi F, de la Rosa E J, Boya P, de Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy. 2012;8:187. doi: 10.4161/auto.8.2.18535. [DOI] [PubMed] [Google Scholar]
- 127.Vellai T, Takács-Vellai K, Sass M, Klionsky DJ. The regulation of aging: does autophagy underlie longevity? Trends Cell Biol. 2009;19:487–494. doi: 10.1016/j.tcb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang C, Liang CC, Bian ZC, Zhu Y, Guan JL. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci. 2013;16:532–542. doi: 10.1038/nn.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang C, Chen S, Yeo S, Karsli-Uzunbas G, White E, Mizushima N, Virgin HW, Guan JL. Elevated p62/SQSTM1 determines the fate of autophagy-deficient neural stem cells by increasing superoxide. J Cell Biol. 2016;212:545–560. doi: 10.1083/jcb.201507023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang C, Haas MA, Yang F, Yeo S, Okamoto T, Chen S, Wen J, Sarma P, Plas DR, Guan JL. Autophagic lipid metabolism sustains mTORC1 activity in TSC-deficient neural stem cells. Nat Metab. 2019a;1:1127–1140. doi: 10.1038/s42255-019-0137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang C, Yang T, Liang M, Xie J, Song N. Astrocyte dysfunction in Parkinson's disease: from the perspectives of transmitted α-synuclein and genetic modulation. Transl Neurodegener. 2021;10:39. doi: 10.1186/s40035-021-00265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang C, Haas M, Yeo SK, Sebti S, Fernández ÁF, Zou Z, Levine B, Guan JL. Enhanced autophagy in Becn1F121A/F121A knockin mice counteracts aging-related neural stem cell exhaustion and dysfunction. Autophagy. 2022;18:409–422. doi: 10.1080/15548627.2021.1936358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang J, Wang Z, Dai L, Zhu X, Guan X, Wang J, Li J, Zhang M, Bai Y, Guo H. Supt16 haploinsufficiency impairs PI3K/AKT/mTOR/autophagy pathway in human pluripotent stem cells derived neural stem cells. Int J Mol Sci. 2023;24:3035. doi: 10.3390/ijms24033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang M, Liang X, Cheng M, Yang L, Liu H, Wang X, Sai N, Zhang X. Homocysteine enhances neural stem cell autophagy in in vivo and in vitro model of ischemic stroke. Cell Death Dis. 2019b;10:561. doi: 10.1038/s41419-019-1798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang S, Li B, Qiao H, Lv X, Liang Q, Shi Z, Xia W, Ji F, Jiao J. Autophagy-related gene Atg5 is essential for astrocyte differentiation in the developing mouse cortex. EMBO Rep. 2014;15:1053–1061. doi: 10.15252/embr.201338343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Watchon M, Luu L, Plenderleith SK, Yuan KC, Laird AS. Autophagy function and benefits of autophagy induction in models of spinocerebellar ataxia type 3. Cells. 2023;12:893. doi: 10.3390/cells12060893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu MY, Lu JH. Autophagy and macrophage functions: inflammatory response and phagocytosis. Cells. 2020:9–70. doi: 10.3390/cells9010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, Diwan A, Lee JM. Enhancing astrocytic lysosome biogenesis facilitates Aβclearance and attenuates amyloid plaque pathogenesis. J Neurosci. 2014;34:9607–9620. doi: 10.1523/JNEUROSCI.3788-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xilouri M, Brekk OR, Landeck N, Pitychoutis PM, Papasilekas T, Papadopoulou-Daifoti Z, Kirik D, Stefanis L. Boosting chaperone-mediated autophagy in vivo mitigates α-synuclein-induced neurodegeneration. Brain. 2013;136:2130–2146. doi: 10.1093/brain/awt131. [DOI] [PubMed] [Google Scholar]
- 140.Xu R, Duan C, Meng Z, Zhao J, He Q, Zhang Q, Gong C, Huang J, Xie Q, Yang Q, Bai Y. Lipid microcapsules promoted neural stem cell survival in the infarcted area of mice with ischemic stroke by inducing autophagy. ACS Biomater Sci Eng. 2022;8:4462–4473. doi: 10.1021/acsbiomaterials.2c00228. [DOI] [PubMed] [Google Scholar]
- 141.Xu Y, Propson NE, Du S, Xiong W, Zheng H. Autophagy deficiency modulates microglial lipid homeostasis and aggravates tau pathology and spreading. Proc Natl Acad Sci USA. 2021;118:e2023418118. doi: 10.1073/pnas.2023418118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xue X, Wang LR, Sato Y, Jiang Y, Berg M, Yang DS, Nixon RA, Liang XJ. Single-walled carbon nanotubes alleviate autophagic/lysosomal defects in primary glia from a mouse model of Alzheimer's disease. Nano Lett. 2014;14:5110–5117. doi: 10.1021/nl501839q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yazdankhah M, Farioli-Vecchioli S, Tonchev AB, Stoykova A, Cecconi F. The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis. 2014;5:e1403. doi: 10.1038/cddis.2014.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yim WW, Mizushima N. Lysosome biology in autophagy. Cell Discov. 2020;6:6. doi: 10.1038/s41421-020-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yin Y, Li E, Sun G, Yan HQ, Foley LM, Andrzejczuk LA, Attarwala IY, Hitchens TK, Kiselyov K, Dixon CE, Sun D. Effects of DHA on hippocampal autophagy and lysosome function after traumatic brain injury. Mol Neurobiol. 2018;55:2454–2470. doi: 10.1007/s12035-017-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zatyka M, Sarkar S, Barrett T. Autophagy in rare (NonLysosomal) neurodegenerative diseases. J Mol Biol. 2020;432:2735–2753. doi: 10.1016/j.jmb.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao L, Biesbroek JM, Shi L, Liu W, Kuijf HJ, Chu WW, Abrigo JM, Lee RK, Leung TW, Lau AY, Biessels GJ, Mok V, Wong A. Strategic infarct location for post-stroke cognitive impairment: a multivariate lesion-symptom mapping study. J Cereb Blood Flow Metab. 2018;38:1299–1311. doi: 10.1177/0271678X17728162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhen Y, Stenmark H. Autophagosome biogenesis. Cells. 2023;12:668. doi: 10.3390/cells12040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhou X, Zhou J, Li X, Guo C, Fang T, Chen Z. GSK-3beta inhibitors suppressed neuroinflammation in rat cortex by activating autophagy in ischemic brain injury. Biochem Biophys Res Commun. 2011;411:271–275. doi: 10.1016/j.bbrc.2011.06.117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


