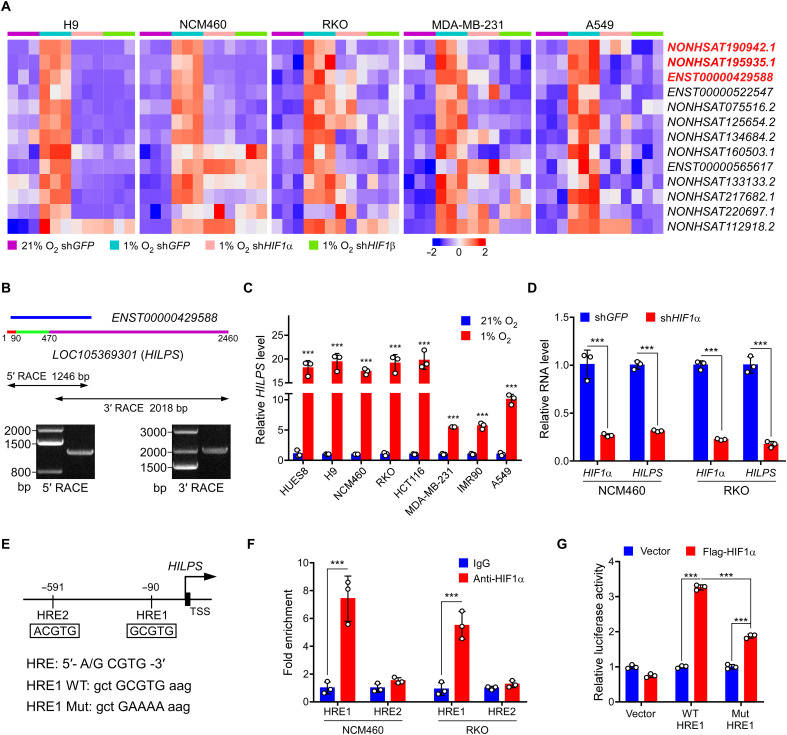
Fig. 1. Characterization of HILPS, a novel human noncoding RNA globally activated by HIF1α at hypoxia.
(A) Heatmap presentation of 13 converged hypoxia-induced lncRNAs in five cell lines (fold change > 1.5, P < 0.05). Expression changes in the presence or absence of HIF1α/HIF1β are presented. Top three candidates are labeled in red. shGFP, control short hairpin RNA (shRNA) targeting GFP. (B) 5′ and 3′ RACE amplicons of HILPS using total RNA isolated from RKO cells exposed to 1% O2 for 24 hours as a template. Schematic of ENST00000429588 and full-length HILPS is illustrated at the top. Three exons of HILPS are marked in colors: exon 1 (1 to 90 nt) in red, exon 2 (91 to 470 nt) in green, and exon 3 (471 to 2460 nt) in purple. 5′ and 3′ RACE PCR amplicons and PCR products are shown in the middle and bottom. (C) qPCR analysis of HILPS in various cell lines exposed to 21% or 1% O2 for 24 hours. (D) qPCR analysis of HILPS in HIF1α-depleted NCM460 and RKO cells exposed to 1% O2 for 24 hours. (E) Schematic illustration of HIF1α binding sites on the HILPS locus. HRE1 consensus sequence mutations are shown as HRE1 mutant (Mut). TSS, transcription start site. (F) ChIP analysis of HIF1α binding to the HILPS locus in NCM460 and RKO cells. IgG, immunoglobulin G. (G) Luciferase reporter assay of the HILPS promoter construct containing HRE1 WT or mutant in the presence or absence of exogenous HIF1α in RKO cells. Data shown are means ± SD from biological triplicates. ***P < 0.001; unpaired two-tailed Student’s t test [(C), (D), and (F)] and one-way analysis of variance (ANOVA) (G).