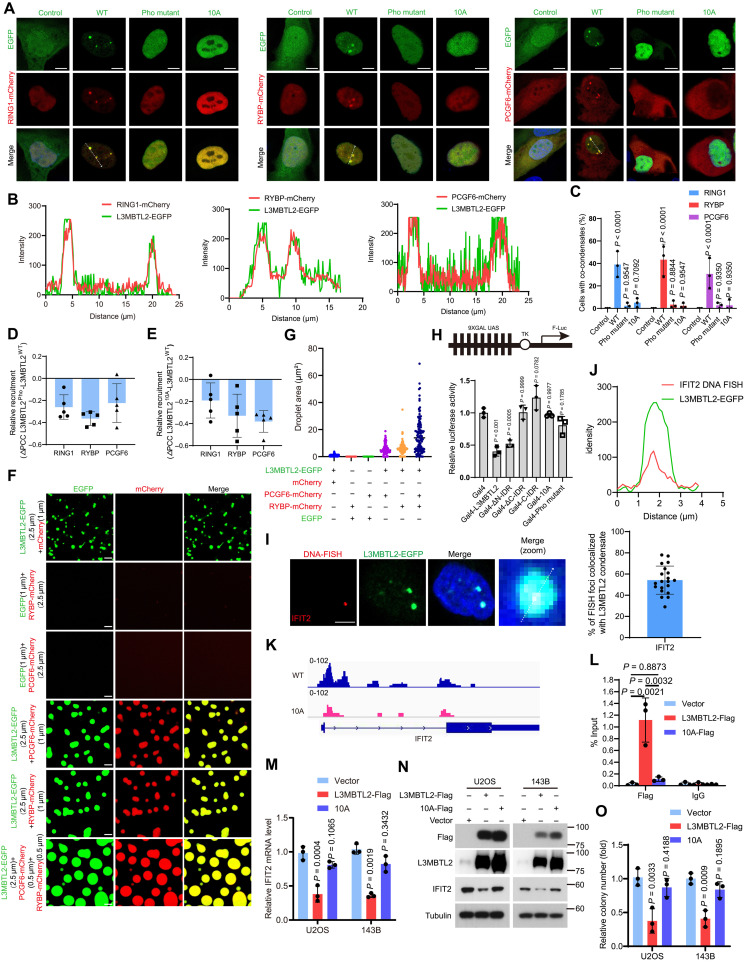
Fig. 4. L3MBTL2 condensates execute its tumor suppression by selectively enriching the components of PRC1.6 complex.
(A) Representative images of U2OS cells cotransfected with the indicated plasmids for 24 hours. Scale bars, 5 μm. (B) Line scan analysis of the fluorescence intensity along with the indicated line in (A). (C) Percentage of n = 100 cells with co-condensates in the indicated cells from (A). (D and E) Relative change in recruitment of the indicated PRC1.6-mCherry for L3MBTL2 wild type, Pho (D), or 10A (E). n = 5. means ± SD. (F) Representative images of the indicated proteins in the phase separation buffer with 25 mM NaCl for 1 min at 25°C. Scale bars, 5 μm. (G) Quantification of the droplets area in (F) was calculated. (H) HEK293T cells were transfected with the indicated constructs, along with the 9xGal-TK-luc reporter and the Renilla control reporter for 24 hours. Then, cells were analyzed for the relative luciferase activity. (I) Left: Confocal images of L3MBTL2-EGFP condensates and IFIT2 loci by DNA FISH in U2OS cells stably expressing pTeton-L3MBTL2-EGFP. Right: Quantification of DNA FISH foci colocalized with L3MBTL2 nuclear condensate (n = 20). Scale bar, 5 μm. (J) Line scan analysis of the fluorescence intensity along with the indicated line in (I). (K) Genome browser screenshots of CUT&Tag tracks illustrating binding of L3MBTL2 wild type and 10A to the IFIT2 promoter in the indicated 143B stable cells. (L) Chip-qPCR analysis of L3MBTL2 occupancy at IFIT2 promoter region in the indicated stable cells. n = 3. means ± SD. (M) The relative IFIT2 mRNA levels were normalized to the GAPDH levels in the indicated stable cells, as determined by RT-qPCR. n = 3. means ± SD. (N) The indicated stable cells were analyzed by Western blotting. (O) Colony formation assays were performed for the indicated stable cells. n = 3. means ± SD. Data in (A, C, F, I, and N) are representative of n = 3 biologically independent experiments.