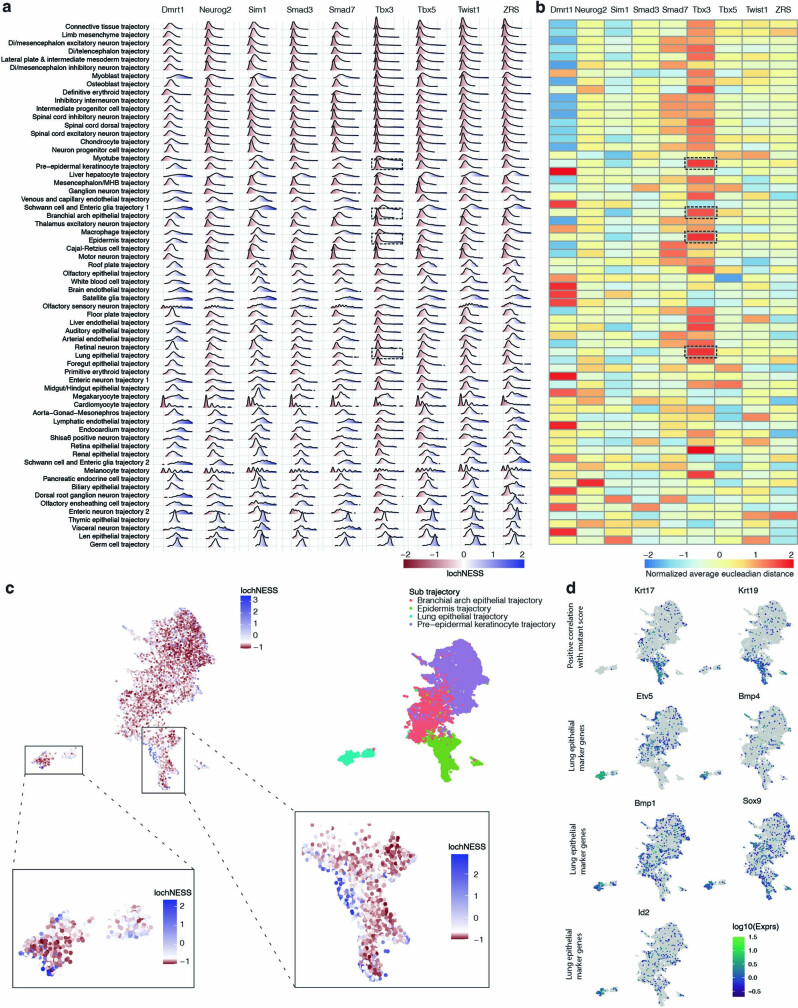
Extended Data Fig. 9. Systematic screening of lochNESS distributions identifies altered epithelial sub-trajectories in the Tbx3 TAD Boundary KO mutant.
a, Distribution of lochNESS in each sub-trajectory of the mutants in the FVB background strain, all of which are TAD boundary KOs. Dashed boxes in the sixth column highlight the most deviated epithelial sub-trajectories in the Tbx3 TAD Boundary KO mutant. b, Row-normalised heatmap showing the average euclidean distance between lochNESS and lochNESS under permutation in each sub-trajectory for the same mutants shown in panel a, centred and scaled by row. Dashed boxes in the sixth column again highlight the most deviated epithelial sub-trajectories in the Tbx3 TAD Boundary KO mutant. c, UMAP showing co-embedding of Tbx3 TAD Boundary KO and pooled wildtype cells in the pre−epidermal keratinocyte, epidermis, branchial arch, and lung epithelial sub-trajectories, coloured by lochNESS (top left) [with blown up insets showing lochNESS in lung epithelial (bottom left) and epidermis (bottom right) sub-trajectories], or by sub-trajectory identity (right). LochNESS colour scale is centred at the median of lochNESS. d, same as in panel c, but coloured by expression of selected mutant related genes and marker genes.