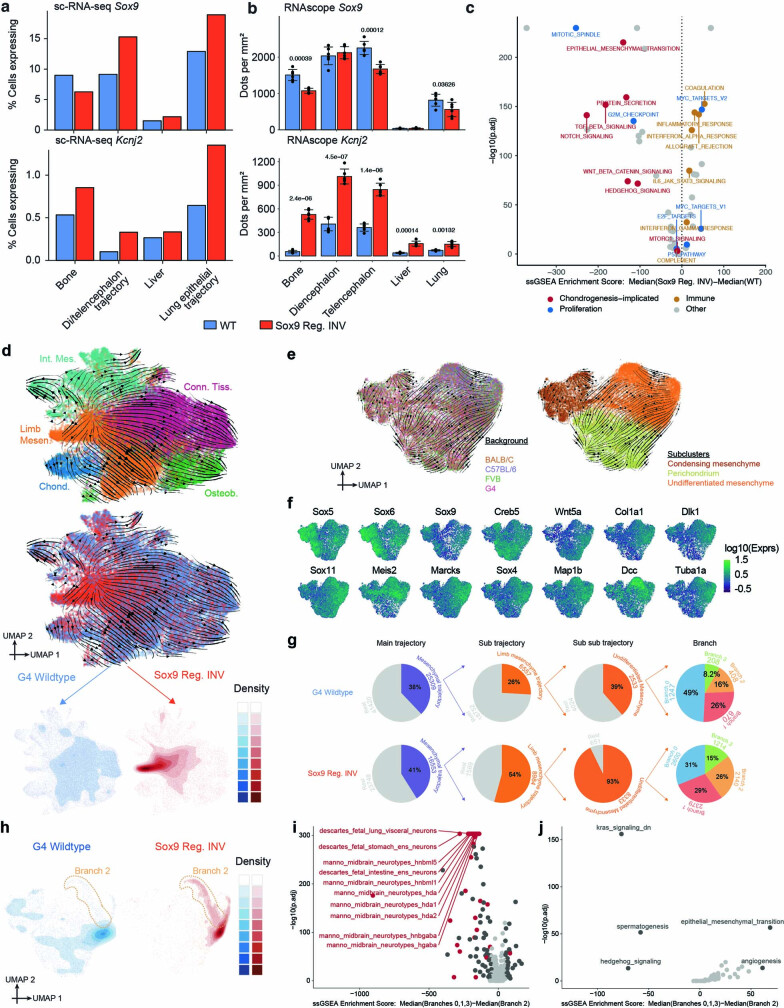
Extended Data Fig. 12. Misregulation of Sox9 and Kcnj2, and stalling of cells in the undifferentiated mesenchyme in the Sox9 regulatory INV mutant.
a, Quantification of Sox9 (top row) and Kcnj2 (bottom row) expression in scRNA-seq data in the wildtype (blue) and Sox9 regulatory INV (red) genotypes in selected trajectories. For “bone” and “liver”, multiple sub-trajectories were pooled to match the tissue labels in the RNAscope data in panel b. Specifically, “bone” refers to cells from chondrocyte, osteoblast, and limb mesenchyme trajectories, whereas “liver” refers to cells from the liver endothelial and liver hepatocyte trajectories. The bars represent singular values represented as a fraction of 21128, 6375, 1728 and 229 cells in Bone, Di/telencephalon trajectory, Liver and Lung epithelial trajectories, respectively. b, Quantification of Sox9 and Kcnj2 expression based on RNAscope images of heterozygous E13.5 wildtype and Sox9 regulatory INV mutant embryos (n = 6 embryos for each condition). The mRNA signal was counted in a defined area (1 × 1 mm²). The numbers represent p-values of the differences between means calculated using a two-sided student t-test. Non-significant p-values (>0.05) not shown. Error bars represent standard deviation. c, Gene set enrichment analysis on bone cells. Comparison of median ssGSEA65 scores between the Sox9 regulatory INV and wild type for Hallmark gene sets66. Gene sets categorised as proliferation and immune-signalling66 highlighted in blue and brown. Gene sets manually identified to be implicated in chondrogenesis highlighted in red. Note: Bone cells include cells from chondrocyte, osteoblast, and limb mesenchyme trajectories. Many of the hallmark pathways downregulated in the mutant are related to proliferation and chondrocyte differentiation (e.g. mitotic spindle, TGF-β signalling, notch signalling, wnt/β-catenin signalling, protein secretion, epithelial-to-mesenchymal transition), which are known to be mediated by Sox9. Additionally, six of the seven immune-related hallmark pathways were upregulated in the mutant, possibly a secondary effect, as to our knowledge Sox9 is not established to be involved in immune signalling. d, RNA velocity of mesenchymal G4 wildtype and Sox9 regulatory INV cells labelled by sub-trajectories (top) or genotype (middle) and the corresponding 2D density plots split by genotype (bottom). e, Sub-clustering of the limb mesenchyme sub-trajectory based on cells from pooled wildtype. RNA velocity arrows generated using scVelo (Methods) indicate the transition of undifferentiated mesenchyme (marked by Meis2, Marcks, Map1b) into perichondrium (Wnt5a,Creb5) and condensing mesenchyme (Sox5, Sox6, Sox9) in all wildtype samples67–72. f, Marker gene expression used to annotate limb mesenchyme sub-clusters. All except Dcc and Tuba1a are literature-based markers of the three cell types. Note: Because the annotation of “limb mesenchyme” sub-trajectory was propagated forward from earlier stages of development during the creation of MOCA, it is possible that other, non-limb mesenchymal populations also contribute to this expanded, undifferentiated pool in the Sox9 regulatory INV embryos. g, Proportion and the number of cells at different levels of clustering, leading up to the four branches of the undifferentiated mesenchyme. h, Density plots for UMAP embedding of G4 wildtype and Sox9 regulatory INV cells in the limb mesenchymal trajectory (same embedding as Fig. 5e). Dotted lines highlight Branch 2 of the undifferentiated mesenchyme, based on the sub-clustering shown in Fig. 5f. Comparison of the ssGSEA65 scores between the two branches of undifferentiated mesenchyme for Sox9 regulatory INV cells for (i) cell type signature (C8) and (j) Hallmark gene sets. Gene sets that are both significantly different between the two branches and that have a difference in median ssGSEA scores greater than 50 are highlighted in dark grey, and the most significantly different gene sets are also labelled. In panel i, all significantly different gene sets with names containing “neuro” are highlighted in red.