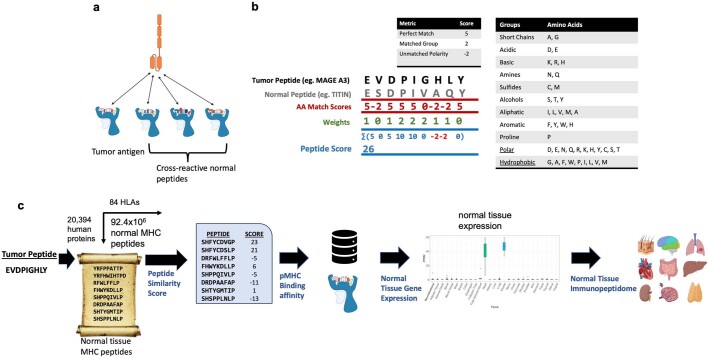
Extended Data Fig. 12. sCRAP cross-reactivity algorithm.
a. Cross-reactivity algorithm was developed to identify peptides presented on normal tissue with similar biophysical properties to tumor antigens such as to pre-emptively predict cross-reactivities and screen for specificity. b. Illustration of peptide scoring system described in methods. c. Schematic of algorithm workflow describing how tumor peptides are scored against each peptide predicted to be presented from the normal proteome (totaling 92.4 × 106 potential MHC peptides). Binding affinity is predicted for each normal peptide and maximum gene expression of parent gene are factored into the overall score of each peptide. Peptides are referenced against a normal tissue immunopeptidomics database. Created with BioRender.com.