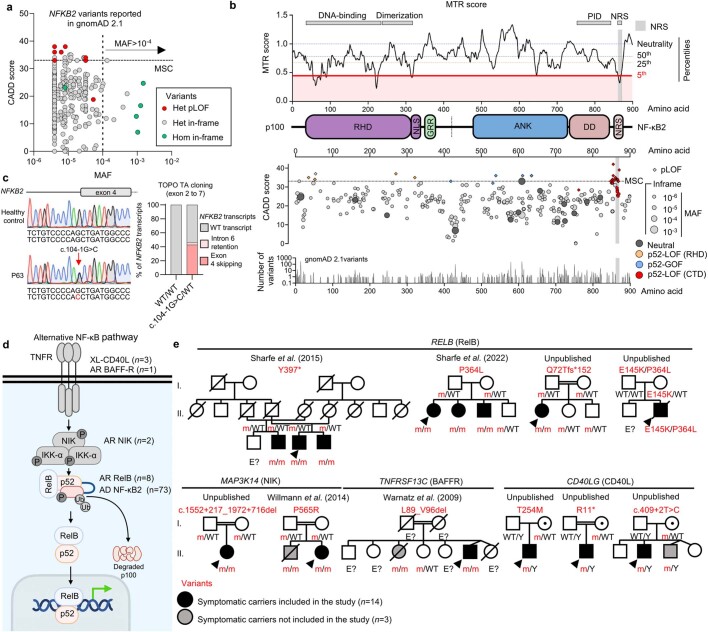
Extended Data Fig. 2. Population genetics and constraint metrics of the NFKB2 gene, and pedigrees of the patients with inborn errors of RelB, NIK, BAFFR and CD40L.
(a) CADD-MAF graph for the NFKB2 variants reported in the gnomAD v2.2.1. The red and grey dots represent monoallelic pLOF and heterozygous in-frame (missense and indel) variants, respectively. The green dots represent homozygous missense variants. The horizontal dashed line represents the mutation significance (MSC) cutoff threshold of 33 for NFKB2. (b) Genomic constrained coding regions across NFKB2, as estimated by the missense tolerance ratio (MTR) score evaluating region-specific intolerance to missense variants. A score <1.0 indicates a lower-than-expected ratio of missense to synonymous variants in the gnomAD v2.0 dataset for the 21 bp window surrounding an amino-acid residue. The horizontal-coloured dashed lines represent the percentiles for the most missense-depleted regions of NFKB2. The NIK-responsive sequence (NRS) is within the 5th percentile for the most missense-depleted regions for NFKB2. The lower graph shows the distribution of the heterozygous NFKB2 variants reported in gnomAD 2.1.1 and from the patients reported in this study, by location within the protein and CADD score. (c) Electropherograms showing the c.104-1 G > C/WT essential splice-site variant carried in the heterozygous state of P63 and a healthy donor (left) and the proportion of transcripts identified by sequencing 100 colonies from TOPO cloning with cDNA from PCR products corresponding to a region spanning exon 2 to 7 in P63 or a healthy donor. (d) Representation of the alternative NF-κB pathway and the patients included. (e) Pedigrees and variants of patients with inborn errors of RelB, NIK, BAFFR and CD40L. A dot within a white symbol indicates an asymptomatic carrier; an arrow indicates the index case; a black diagonal line indicates a deceased individual. “E?” indicates individuals of unknown genotype.