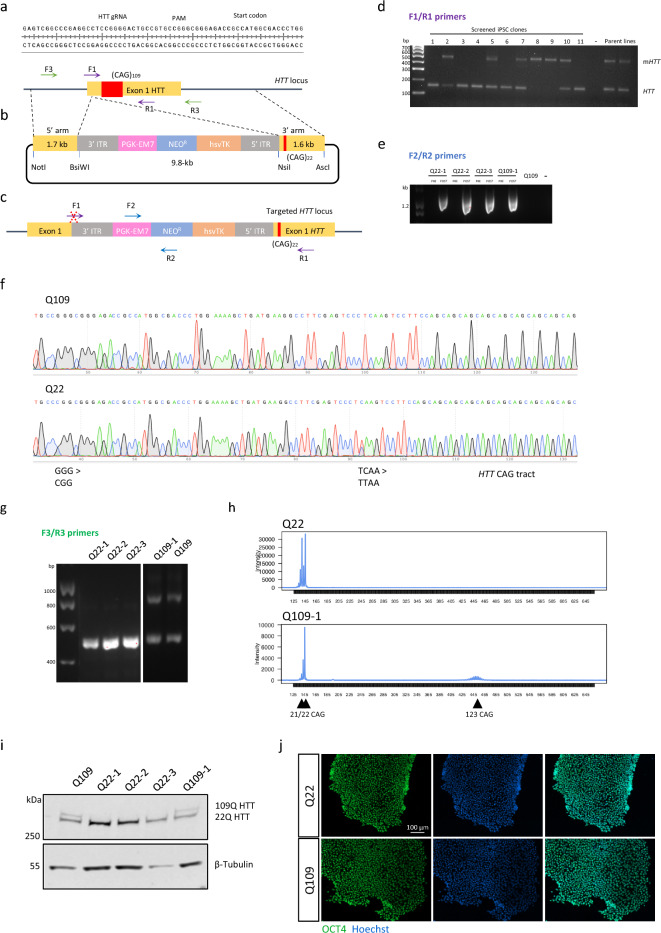
Figure 1.
Design and generation of isogenic Q22 iPSC using piggyBac and CRISPR-Cas9 mediated homologous recombination. (a) Schematic showing HTT sequence and CRISPR-Cas9 guide RNA target site. (b) Schematic depicting the donor DNA and piggyBac transposon-based selection strategy used to target the CAG tract in exon 1 of the HTT locus. The 5′ and 3′ homology arms of the plasmid are 1.7 kb and 1.6 kb, respectively. (c) Schematic of targeted HTT locus and indication of forward and reverse (F/R) primers used for genotyping in panels (d) and (e). (d) Diagnostic PCR screen amplifying exon 1 of HTT locus using F1/R1 primers. Insertion of the targeting repair vector disrupted the forward primer binding site, and thus no PCR amplification was detected in the alleles that were successfully targeted. PCR identified clones in which the WT allele or the mutant allele had been targeted. (e) PCR screen amplifying the selection cassette illustrates loss of selection cassette in selected clones after excision. (f) Sanger sequencing of exon 1 of the HTT locus using F3/R3 primers confirmed excision of the piggyBac cassette from TTAA integration sites and introduction of silent PAM site mutations (GGG > CCG). (g) PCR amplifying exon 1 of the HTT locus in selected clones verified successful correction at the HTT locus. (h) Electropherograms of fluorescent PCR across the CAG repeat showed two WT alleles (21 CAG, 22 CAG) present and loss of the expanded allele in Q22 iPSC. Clone Q109-1 retained the expanded repeat, and the WT allele is re-inserted with a CAG repeat length + 1 of original WT allele. (i) Western blot showing expression of wildtype size HTT (22Q) following cassette excision, confirming successful correction at the HTT locus in selected clones. (j) Gene-corrected clones maintained expression of the pluripotency marker OCT4, shown by immunostaining. Scale bar = 100 μm. Uncropped gels and plots are presented in Supplementary Fig. S5.