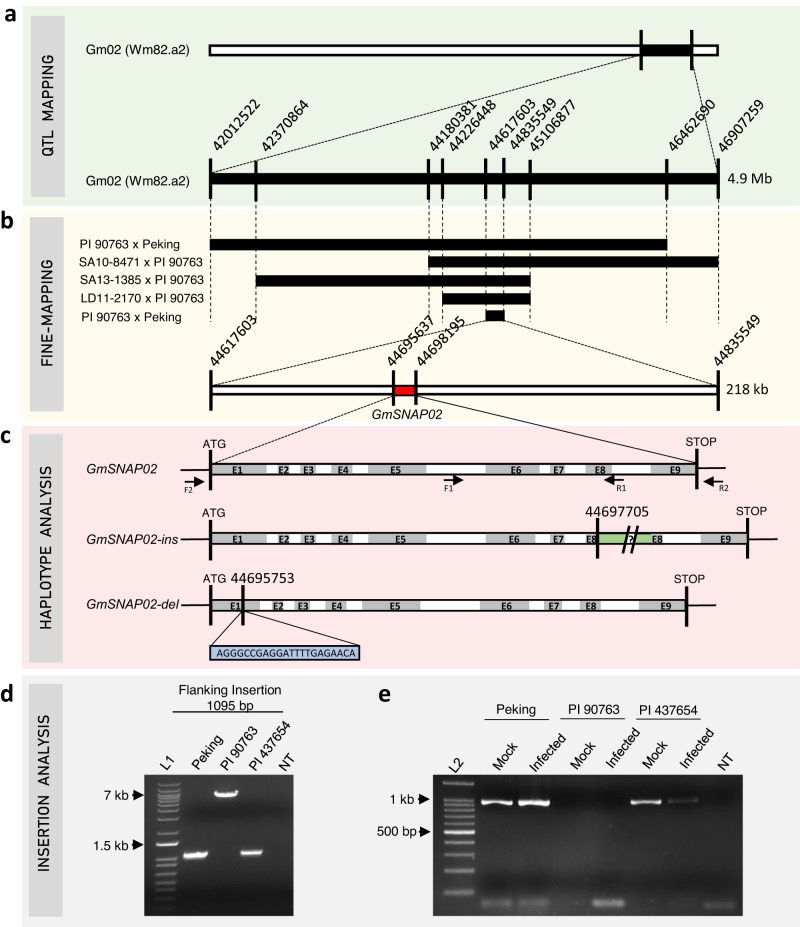
Fig. 2. GmSNAP02 positional cloning.
a The QTL02 confidence interval Gm02: 42,012,522–46,907,259 (Wm82.a2) was identified from a cross between PI 90763 × Peking. b An initial fine-mapped region of ~880 kb containing 112 genes was determined using recombinant inbred F3:4 lines derived from four populations. F4:5 lines derived from the cross between PI 90763 x Peking were used to further narrow the region to ~218 kb containing 34 genes. Within this region GmSNAP02 (Glyma.02G260400) became a candidate gene (red). c Haplotypes identified at GmSNAP02 included the susceptible GmSNAP02 haplotype of the Williams 82 reference genome, GmSNAP02-ins, a resistant haplotype caused by an ~6 kb insertion (green) in exon 8 in PI 90763, and GmSNAP02-del haplotype caused by a 22 nt deletion (blue) in exon 1 resulting in a frameshift mutation leading to a premature stop. GmSNAP02 gene-specific primers designed to flank the insertion in exon 8 (d, F1/R2) amplified products of the predicted size in Peking and PI 437654, but a larger product in PI 90763, confirming the presence of an insertion. This experiment was repeated four times with similar results. e GmSNAP02 gene-specific primers designed immediately upstream of the start and downstream of the stop codons (c, F2/R2) amplified a full-length GmSNAP02 cDNA sequence from mock-inoculated and SCN-infected (3 days post inoculation) roots of Peking and PI 437654, but not from PI 90763. Sequencing of the product amplified from PI 437654 confirmed the presence of a 22 nt deletion in exon 1 of GmSNAP02. This experiment was repeated twice with similar results. L1 = 1 kb Plus ladder (Invitrogen), L2 = 100 bp ladder (NEB), NT = no template control.