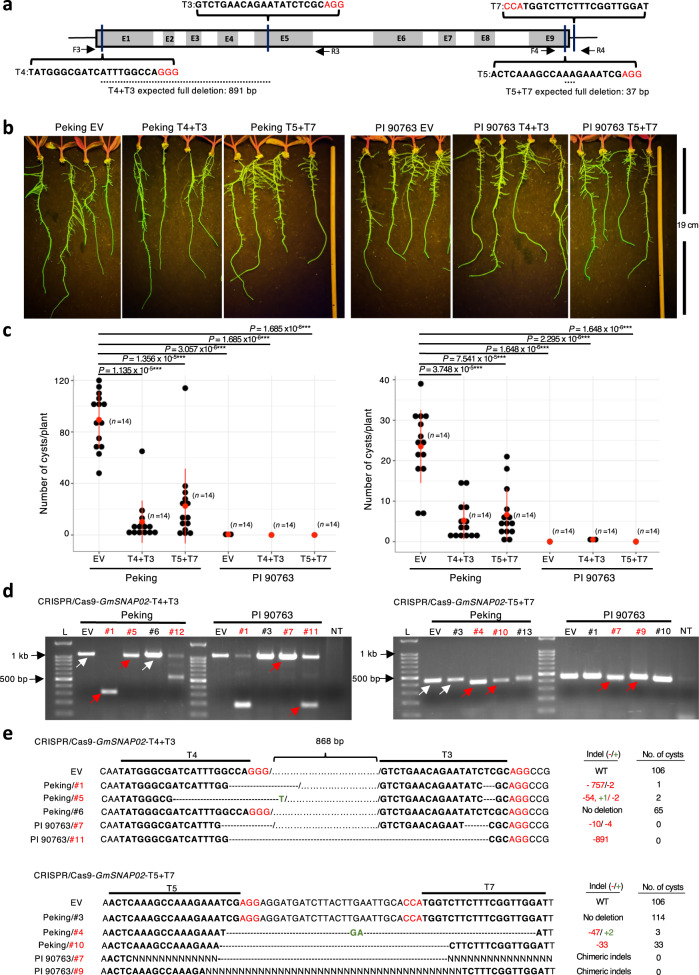
Fig. 4. Functional validation of GmSNAP02 in resistance to SCN using CRISPR/Cas9.
a Diagram showing the positions of the guide RNA (gRNA) sequences designed to edit the GmSNAP02 gene. PAM sequences are in red. gRNA sequences are bolded. b Composite soybean plants with transgenic GFP-positive hairy roots were selected under fluorescent light. Representative images are shown. No gross phenotypic differences were observed in GmSNAP02-edited roots of either Peking or PI 90763 (n = 14 plants/construct examined in two independent experiments). Pictures taken just before transplanting and nematode inoculation. c Cyst counts on transgenic roots of Peking and PI 90763 composite plants transformed with K599 carrying empty vector (EV; control), CRISPR/Cas9-GmSNAP02-T4 + T3, and CRISPR/Cas9-GmSNAP02-T5 + T7 constructs, respectively. Data are shown for two biological replicates for each construct and genotype. Means ± s.d. are denoted with a red dot and line (n = 14). ***P < 0.001 and **P < 0.01, Wilcoxon rank-sum statistical test. d Amplified fragments from genomic DNA extracted from roots transformed with CRISPR/Cas9-GmSNAP02-T4 + T3, and CRISPR/Cas9-GmSNAP02-T5 + T7, respectively. Fragments flanking T4 and T3 gRNA cleavage sites were amplified using F5 and R5 primers (shown in (a)). Fragments flanking T5 and T7 gRNA cleavage sites were amplified using F6 and R6 primers (shown in (a)). A subset of roots from both genotypes for each construct was selected for genotyping. Fragments indicated by the arrows were gel extracted, subcloned, and sequenced to confirm deletions. White arrows no deletion, Red arrows/#’s deletion confirmed, L 100 bp ladder (NEB), NT no template control. e Sequences of selected fragments from d. The number of nucleotides deleted (red font) and/or inserted (green font) and the corresponding cyst counts for each plant are indicated in the columns on the right.