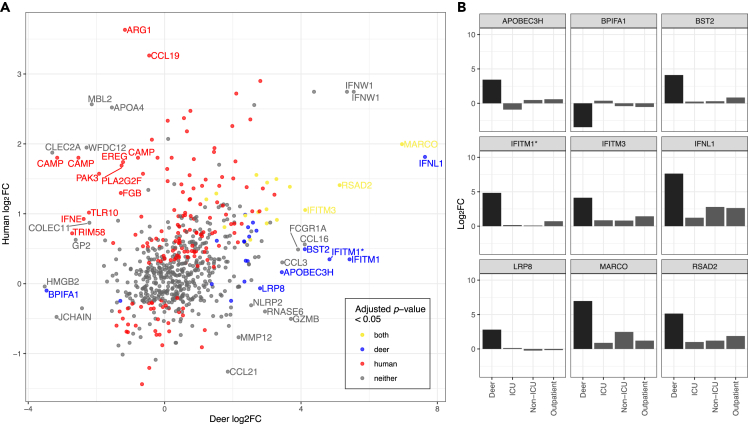
Figure 5.
Comparison of the white-tailed deer (WTD) versus human host response to SARS-CoV-2 infection
(A) The WTD expression fold change is plotted here against the human expression fold change from comparable human samples based on the results of an orthology analysis between WTD and human. The genes are labeled with the respective human gene name, if the absolute difference in log2 fold change values between the human and WTD expression profiles is greater than 2.5. Due to duplicated genes in the WTD genome, some points are labeled with the same human gene name. From left to right, cathelicidin antimicrobial peptide (CAMP) genes are associated with LOC110152352, LOC110152353, and LOC110152344 in the WTD genome, interferon omega 1 (IFNω1) genes are associated with LOC110144825 and LOC110144833 and interferon induced transmembrane protein 1 (IFITM1) genes are associated with LOC110149600 and LOC110149612. Points are colored based on significance in their respective differentially gene expression analysis. Genes with very low expression values are not given a p value in the DESeq2 analysis and are not considered significant.
(B) Genes that are significantly differentially expressed in the WTD SARS-CoV-2 infection and have an absolute difference in log2 fold change values between the human and WTD infections greater than 2.5 are plotted here with their corresponding log2 fold change values across different levels of human disease severity. Intensive care unit (ICU), non-ICU, and outpatient are comparisons of human nasopharyngeal swabs between patients from these respective settings, and healthy patients. Genes are labeled with their human ortholog gene name. IFITM1 here corresponds to LOC110149600 in the WTD genome. See also Table S8.