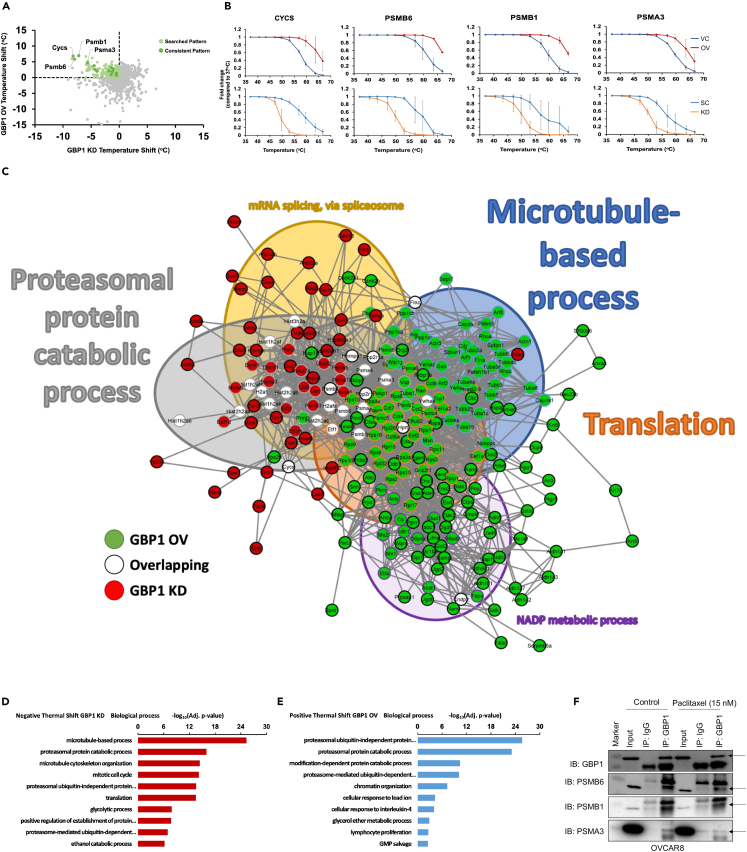
Figure 3.
GBP1 crosstalk identification
Differential thermal proteome profile of ID8 cells stably transduced with GBP2b overexpression (OV) or vector control (VC) or scrambled control (SC) or GBP2b shRNA (KD). Cells were collected and 106 cells in each PCR tube were incubated at different temperatures (37°C, 41°C, 44°C, 47°C, 50°C, 53°C, 56°C, 59°C, 63°C, or 67°C). Cells were lysed and an equal quantity of soluble protein was labeled with tandem mass tag (TMT), followed by liquid chromatography-tandem mass spectrometry analysis. Thermal stabilities were calculated, and thermal stability shifts in GBP2b presence or absence were calculated.
(A) Dot plot for thermal stability shift of proteins in GBP1 OV and KD groups (normalized with vector/scrambled control).
(B) The melting curves for PSMB1, PSMB6, PSMA3, and CYCS indicate a shift in melting temperature in ID8 cells with GBP2b overexpression or knockdown condition compared to vector/scrambled control.
(C) Significant proteins in common across conditions were graphed in String and showed enrichment for the proteasomal protein catabolic process (gray-filled circles), microtubule-based process (blue-filled circles), translation, NADP metabolic process, and mRNA splicing via spliceosome. The thickness of network edges indicates the strength of data supporting the interaction.
(D and E) Gene ontology enrichment analysis for different biological processes.
(D) Gene ontology of proteins with a negative thermal shift by GBP2b-KD.
(E) Gene ontology of proteins with a positive thermal shift by GBP2b-OV.
(F) Immunoprecipitation with anti-GBP1 or anti-IgG antibodies from whole-cell lysate of OVCAR8 cells, followed by western blot for GBP1, PSMB1, PSMB6, and PSMA3.