Abstract

Surface-enhanced Raman scattering (SERS) is a highly sensitive technique used in diverse biomedical applications including rapid antibiotic susceptibility testing (AST). However, signal fluctuation in SERS, particularly the widespread of signals measured from different batches of SERS substrates, compromises its reliability and introduces potential errors in SERS-AST. In this study, we investigate the use of purine as an internal standard (IS) to recalibrate SERS signals and quantify the concentrations of two important purine derivatives, adenine and hypoxanthine, which are the most important biomarkers used in SERS-AST. Our findings demonstrate that purine IS effectively mitigates SERS signal fluctuations and enables accurate prediction of adenine and hypoxanthine concentrations across a wide range (5 orders of magnitude). Calibrations with purine as an IS outperform those without, exhibiting a 10-fold increase in predictive accuracy. Additionally, the calibration curve obtained from the first batch of SERS substrates remains effective for 64 additional substrates fabricated over a half-year period. Measurements of adenine and hypoxanthine concentrations in bacterial supernatants using SERS with purine IS closely align with the liquid chromatography–mass spectrometry results. The use of purine as an IS offers a simple and robust platform to enhance the speed and accuracy of SERS-AST, while also paving the way for in situ SERS quantification of purine derivatives released by bacteria under various stress conditions.
Introduction
Surface-enhanced Raman spectroscopy (SERS) is a sensitive optical sensing method and can rapidly afford an informative spectrum from trace molecules. Based on these features, SERS can be applied in many fields such as toxic compound detection1,2 and biomedical applications.3,4 One developing biological application of SERS with realistic potential is conducting antibiotic susceptibility testing (AST) of infecting bacteria.5−9 Bacterial drug resistance is a global crisis, resulting in a large number of deaths and enormous cost to prevent it and threat the infected patients.10 SERS can provide rapid AST results with high accuracy, and this could not only save lives but also reduce misuse or abuse of antibiotics, leading to reducing the possibility of bacteria for developing the resistance. It has been shown that the majority of SERS spectra from bacteria are primarily composed of purine derivative metabolites, with adenine and hypoxanthine being the main contributors.11,12 These two compounds serve as the biomarkers utilized in SERS-AST, in which the intensities of the SERS breathing modes of adenine and hypoxanthine, released from bacteria treated with varying concentrations of antibiotics, are compared to determine the minimum inhibitory concentration (MIC) and perform AST (for more details, refer to the Supporting Information).8,9 Thus, acquiring stable and reliable SERS signals of adenine and hypoxanthine is crucial for SERS-AST and other bacterial applications of SERS.
SERS amplifies weak normal Raman signals, often reaching enhancement factors larger than 106,13 enabling single-molecule detection in certain cases.14 However, SERS is plagued by its orders-of-magnitude signal fluctuation and low reproducibility.15 Fluctuations arise from variations in the electromagnetic and chemical enhancements. The former originates from the plasmon resonance of nanostructures employed for generating the SERS effect, while the latter originates from the charge transfer between the analyte molecules adsorbed on nanostructures. The plasmon resonance depends on the size, shape, and geometric arrangement of the nanostructures. When the gap between two adjacent plasmonic nanostructures decreases from dozens of nanometers to subnanometer, the so-called “hot spot” effect is created. It boosts the electromagnetic field exponentially16−18 and contributes a large portion of the signal.19,20 Thus, subtle geometry changes of the nanostructures could greatly alter the electromagnetic field, resulting in large signal fluctuations. The surface chemistry of the SERS substrate affects not only the chemical enhancement but also the adsorption behavior of analyte molecules, which must be very close to or adsorb on the substrate surface to receive the electromagnetic and chemical enhancements. Therefore, sample preparation procedure and experimental environments could also affect the stability of the analytes’ SERS signals.21,22 Owing to the lack of atomically precise control over the physical size/geometry and surface chemistry of SERS substrates, most SERS measurements exhibit large fluctuation from point to point (low uniformity) and from batch to batch (low reproducibility). These limitations have prevented SERS from being a widely employed analytical method and also slowed the SERS-AST protocol significantly.
Efforts have been undertaken to mitigate signal fluctuation, particularly due to electromagnetic enhancement. One approach involves enhancing the uniformity and reproducibility of substrates, specifically in the precise control of narrow gaps where a significant portion of SERS signals originate from adsorbed analyte molecules. Traditional top-down methods for solid substrates have been used to control nanostructure geometry, but are often time-consuming, costly, and not easily scalable, thus limiting broader applications.23 In recent years, more cost-effective and scalable top-down techniques have emerged for precise engineering of both in-plane24,25 and out-of-plane plasmonic hot spots.26−28 While these substrates exhibit high uniformity (minimal point-to-point fluctuation), their reproducibility (fluctuation across substrates or batches), which is crucial for practical applications, remains an area requiring further verification. It is worth noting that these methods do not inherently reduce fluctuation stemming from chemical enhancement.
For colloidal particles, molecule linkers have been exploited to precisely control the aggregation of nanoparticles and reduce the gaps among them;21,29,30 as a result, a relative standard deviation (RSD) value below 1% was reported.21 This method could control large amounts of nanoparticles with narrow gaps at once, and the surface is relatively controlled. However, the linker molecules are located in the middle of the gap where the electromagnetic field is enhanced the most, resulting in higher SERS signals of the linker and lower signals of analytes.29 Furthermore, the sizes, shapes, and densities of the nanoparticles on a substrate or in a solution need to be carefully controlled to reduce the fluctuation.21 To maintain reproducibility of the substrate surface and the substrate–analyte interaction, substrate fabrication and storage, sample preparation, and spectrum measurement protocols must be kept consistent. Other preparation procedures such as spin coating or long-time soaking were employed to decrease fluctuations with some degree of success, but the lengthy sample preparation procedure remains an issue.22 Overall, the above approaches are either too costly or time-consuming, and therefore simple/inexpensive SERS quantification methods are necessary to turn SERS into a practical analytical method for detecting trace analytes.
Adding a standard is an effective and well-accepted method for calibrating fluctuating signals in analytical chemistry. External standard methods, conducted by acquiring standard molecules’ SERS signals near the site of an analyte, are a simple way to get reference signals. Unfortunately, it cannot eliminate the point-to-point fluctuation and usually leads to doubling of the time and labor for the analysis. Alternatively, the internal standard (IS) method is conducted by analyzing standards and analytes simultaneously and using the standard’s signals to rescale the analyte’s signals.
Various internal standards (ISs) have been employed for SERS quantification in different applications. These include isotope-edited analytes,31 graphene on PMMA-supported Au nanoparticles,32 or sandwiched molecules in core–shell colloidal nanorods.33 An emerging IS is the pseudopeak derived from plasmon-enhanced electronic Raman scattering (ERS).27,34,35 This pseudopeak holds promise for calibrating incident electric field fluctuations and electromagnetic enhancement, given its analogous local electromagnetic field dependence with analyte SERS signals. Moreover, it is convenient to use since it is inherently present, eliminating the need for additional potentially disruptive chemicals. However, this IS may face challenges when substrate surfaces are affected by ambient air or when analyte solution composition fluctuates under practical conditions. Furthermore, the pseudopeak typically falls within the <200 cm–1 region, which is not covered by most commercial Raman systems with fixed gratings. Thus, alternative IS options remain necessary. Using additional chemicals as IS offers flexibility in choosing cost-effective options36 and facilitates the simultaneous calibration of both electromagnetic and chemical enhancements, provided the IS molecule is selected carefully and the calibration workflow (see Figure 1) is followed meticulously. Furthermore, it is crucial to choose an additional chemical IS carefully to avoid overlap with the SERS signals of interest from analytes and to minimize the suppression of their intensities due to adsorption competition.35
Figure 1.
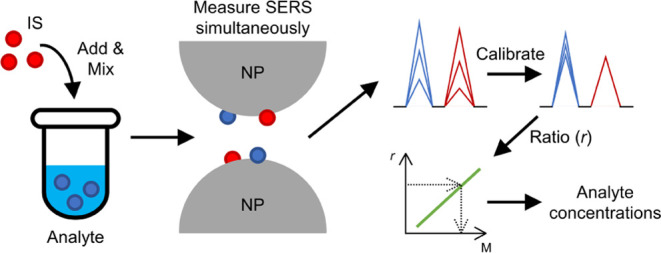
Schematic illustrating the workflow for utilizing a suitable molecule as a SERS IS. The IS and analyte molecules experience similar electromagnetic and chemical enhancements from nanoparticles. The ratio of SERS signals from analytes to IS is used to establish a calibration curve, enabling the measurement of an analyte’s unknown concentration based on signal ratios. Measurements can be taken from various spots on different SERS substrate batches with reduced fluctuations and improved accuracy.
With these considerations in mind, there remains a gap in existing research regarding the optimal IS for the quantification of adenine and hypoxanthine. While some studies have attempted to calibrate the SERS signal for adenine,35,37,38 none have explored the use of ISs for hypoxanthine. Furthermore, the ISs employed in prior works were mostly imbedded into the SERS substrates during fabrication processes, thereby failing to leverage the full benefits of using additional chemicals as IS, such as flexibility in choice and calibration of both electromagnetic and chemical enhancements. Therefore, this study aims to identify an appropriate IS to reduce signal fluctuation and enable the rapid quantification of adenine and hypoxanthine, major SERS peaks of molecules released by bacteria under various challenging conditions, which are vital for applications of SERS to bacteriology.
Purine, the base of purine derivatives, exhibits similar SERS behavior, and it is readily available from various suppliers. More importantly, it has not been identified in the SERS spectra of bacteria11,12 and not been detected in intracellular metabolites of Staphylococcus aureus(39) and Escherichia coli(40) and, more generally, “purine itself has never been found in nature”,41 suggesting that it could be a convenient and accessible IS for calibrating the SERS of other purine derivatives. In this study, we examined the feasibility of purine as an IS to calibrate adenine’s and hypoxanthine’s fluctuating SERS signals and to predict their concentrations in the supernatants of S. aureus and E. coli, respectively. We showed that the use of purine as an IS results in highly accurate predictions of analyte concentrations across 5 orders of magnitude; and leads to precise calibration of the SERS signals of analytes across dozens of substrates spanning several months. Notably, the predicted concentrations of adenine and hypoxanthine in bacterial supernatants align with measurements obtained using liquid chromatography plus mass spectrometry (LC-MS).
Materials and Methods
Chemicals and Materials
Chromium(VI) oxide (99.5%), silver nitrate (≥99.85%), magnesium sulfate heptahydrate (99.5%), and adenine (99%) were purchased from ACROS Organics. Purine (98%), hypoxanthine (≥99%), and formic acid (≥ 99.9%) were purchased from Sigma-Aldrich. Methanol (≥99.9%) and acetone (≥99.9%) were purchased from Honeywell. Sodium hydroxide was purchased from Fisher Scientific. Sulfuric acid (95.0–98.0%), hydrochloric acid (36.5–38.0%), and acetonitrile (≥99.9%) were purchased from J. T. Baker. Ortho-phosphoric acid (85%) was purchased from Scharlau. Mueller–Hinton broth (MHB) was purchased from Merck. All solutions employed in this work were prepared with double-distilled and deionized water (dd-diH2O, A4DL, Lotun & Synergy UV, Merck KGaA, 18.2 MΩ·cm) and used without further purification.
Bacterial Supernatant Preparation
S. aureus ATCC 25213 and ATCC 25923 were obtained from the American Type Culture Collection (ATCC). E. coli BW25113 and DH5α were obtained from the National Institute of Genetics in Japan42 and Protech Technology Enterprise Co., respectively. All bacteria were cultured in MHB at 37 °C for 16–18 h; then, subculturing started under the same conditions. Once the optical density of the bacteria at 600 nm approximately reached 0.5, 1 mL of the bacterial suspensions was washed three times by centrifuging (16,160g, 2 min), replacing 0.9 mL of the supernatants with the same volume of dd-diH2O and subsequently resuspending the pellets. The washed and resuspended bacterial samples were incubated at 37 °C for 30 min to allow the bacteria to release the purines. Finally, 0.5 mL of bacterial supernatants was acquired by centrifuging (16,160g, 10 min) and used for SERS and LC-MS analysis. The entire protocol is illustrated in Figure S5.
AgNP/AAO SERS Substrate Fabrication
The fabrication of AgNP/AAO substrates has been reported previously.16 Briefly, a glass slide was cleaned under ultrasonication for 30 min in the sequence of acetone, a 0.5 N sodium hydroxide solution, dd-diH2O, and methanol. After the cleaned slide was coated with an ∼200 nm aluminum film by sputtering, an anodic aluminum oxide (AAO) pore array was developed on it in a 0.3 M sulfuric acid solution under a 21 V DC bias at 3 °C. Then, the pores were further etched to enlarge the pore diameters in a mixed solution containing 3% chromic acid and 5% ortho-phosphoric acid at 35 °C. Next, silver nanoparticles (AgNPs) were electrochemically deposited into the pores under a 9 V AC bias in a mixed solution containing 0.1% silver nitrate and 4.1% magnesium sulfate at 31 °C. After the NPs reached the desired size, the whole slide was withdrawn from the solution, immersed in a 0.2 N hydrochloric acid solution, and rinsed with dd-diH2O. Subsequently, the slide was dried entirely under N2 gas flow and stored individually in a vacuum-sealed plastic bag. The final product is a two-dimensional hexagonal-packed AgNP array with an average diameter of 50 nm, a mean gap of ∼10 nm between neighboring AgNPs, and a SERS-active area of 2.5 cm × 5.5 cm (Figure S2).
Raman Instrumentation and SERS Measurement
SERS spectra were acquired with a typical Raman microscope system. The system integrated a standard upright microscope (BX61WI, Olympus) with a Raman fiber optic probe (SuperHead, Horiba) and a spectrometer (HE 633, Horiba) attached by a thermoelectrically cooled charge-coupled device (CCD) (354308, Horiba). An exciting light beam emitted by a 632.8 nm He–Ne laser (LGK 7665 P18, Lasos) was first delivered via an optical fiber to a laser-line filter to remove unwanted light in the Raman probe before being focused on the surface of the SERS substrate by a 20× objective (MPlanFL N, Olympus, laser spot size: ∼25 μm). The laser irradiation power was 0.5 mW. The scattered light was collected backward with the same objective and filtered by a long-pass filter to remove the elastically scattered light. Then, the light was delivered through an optical fiber to the spectrometer and the CCD for spectral recording. The resultant spectral resolution and error were 3–4 and 0.1 cm–1, respectively. To acquire SERS spectra, 1 μL of solutions was dropped onto the substrates, and at least 10 spectra of each solution were collected around the droplet’s center with a 40 μm spacing. All SERS measurements began 1 min after the dropping and ended within 1 min. The integration time of a spectrum was 1 s, and each spectrum measurement was averaged over 3 repeat cycles. Occasionally, some spectra with extremely dissimilar SERS patterns were discarded.
LC-MS
Liquid chromatography plus mass spectrometry was performed on an ultraperformance liquid chromatography system (Waters ACQUITY UPLC system, Waters), coupled with a hybrid ion trap-orbitrap mass spectrometer (Orbitrap Elite, Thermo Fisher Scientific). Chromatographic separation of a 10 μL sample was conducted with a reversed-phase C18 column (ACQUITY UPLC HSS T3, Waters; particle size: 1.8 μm, inner diameter: 2.1 mm, length: 150 mm, pore size: 10 nm) maintained at 40 °C. The mobile phase consisted of 2% (v/v) acetonitrile and 0.1% (v/v) formic acid in deionized water (A) and 0.1% (v/v) formic acid in acetonitrile (B) at a flow rate of 0.4 mL/min with the following linear gradient: 0–3 min, 99.5–75% A; 3–4.5 min, 75–25% A; 4.5–4.6 min, 25–99.5% A; and 4.6 to 6 min, 99.5% A. The positive mode of electrospray ionization was employed, and the scan range was between 50 and 800 m/z with a resolution of 60000.
Data Processing
The continuum background of a SERS spectrum was estimated and subtracted by a nonlinear iterative peak clipping algorithm.43 Because a contamination peak occasionally appears at 745 cm–1, 3 Gaussian peak functions were utilized simultaneously to fit the 705–838 cm–1 wavenumber range and the peak areas of hypoxanthine or adenine, contaminant, and purine were found. The processing above was conducted by Python 3 along with the third-party libraries Numpy and Scipy. The LC-MS data analysis was conducted on Xcalibur (Thermo Fisher Scientific). Chromatograms of target m/z were obtained with a 10 ppm tolerance, and peaks of adenine and hypoxanthine standards, as well as the bacterial supernatants at the same retention time as the standards, were integrated. Calibration curves were derived by linearly interpolating SERS with purine and LC-MS and by linearly interpolating and extrapolating SERS without purine.
Concentration Prediction
Concentrations of adenine and hypoxanthine were predicted by LC-MS and SERS with or without purine as the IS. In the LC-MS analysis, pure adenine and hypoxanthine solutions were used as external standards (Figure S6). Bacterial supernatants were analyzed immediately after the standard solutions to prevent long-term drift and ensure accurate measurement of adenine and hypoxanthine concentrations. These concentrations in the bacterial supernatants served as references for evaluating the accuracy of SERS predictions. For SERS, construction of calibration curves and predictions of concentrations were performed on different SERS substrates to simulate real-world scenarios. For predictions by SERS without purine IS, droplets of the solutions or supernatants to be tested were directly dropped onto SERS substrates while in SERS with purine, solutions or supernatants to be tested were mixed with purine solutions (volume ratio = 1:1) to match the concentration used in the calibration curves. For pure adenine or hypoxanthine, predictions were based on the corresponding calibration curves, while bacterial supernatant predictions were initially made using the most likely concentration order. If a peak ratio fell outside the range of the calibration curves, lower- or higher-concentration orders were used accordingly. Unusable ratios were excluded from predictions. In cases where multiple curves provided predictions, the concentrations were averaged for the final prediction. The complete concentration prediction procedure for samples with unknown concentrations is illustrated in Figure S7.
Results and Discussion
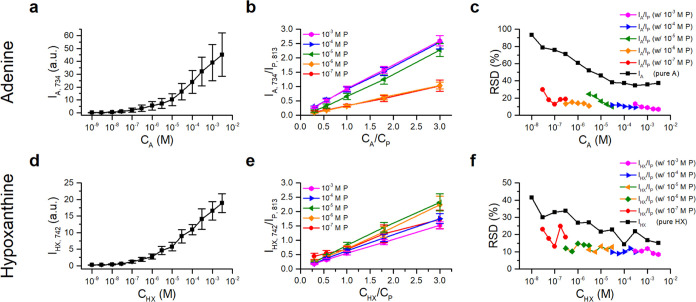
Purine derivatives typically display a strong peak in the 500–850 cm–1 range, attributed to the breathing mode of the purine rings, which has been widely used for analysis in various studies.7−9,44,45Figure S3 shows typical SERS spectra of hypoxanthine, adenine, and purine with the highest peaks observed at 742 cm–1 for hypoxanthine and 734 cm–1 for adenine, respectively. Purine exhibits a distinct peak at 813 cm–1 well separated from the peaks of adenine and hypoxanthine; and can be clearly separated by a low-resolution spectrometer commonly used in many real-world applications, with a resolution of approximately 20 cm–1. In contrast, the difference in the breathing mode frequency between a purine derivative and its isotopologue, which is often unavailable or expensive, is typically smaller than ∼20 cm–1. Therefore, the 813 cm–1 peak of purine serves as a convenient and accessible IS signal for calibrating the corresponding breathing mode signals of hypoxanthine and adenine. Furthermore, given that adenine and hypoxanthine are both purine derivatives with minor functional group substitutions, they, along with purine, are expected to exhibit similar physical and chemical properties, as well as adsorption behavior on a SERS substrate.
Competitive adsorption between analytes and IS molecules could result in suppression of either the analyte or IS signal, leading to improper signal ratios and an inadequate calibration curve.31 To obtain effective calibration curves for analyte prediction across various concentration ranges, it is crucial to identify the appropriate purine/adenine and purine/hypoxanthine ratios. As shown in Figure 2a,2b, respectively, the ring-breathing mode signal of adenine and hypoxanthine decreases as the purine concentration increases. Clear signals of both analytes are observed only when the concentration ratio of purine to adenine (or hypoxanthine) falls within the ranges 10 and 1/10. This indicates similar Raman cross-section and adsorption behavior on the SERS substrates.
Figure 2.
SERS spectra of adenine and hypoxanthine mixed with purine IS at various concentration ratios (a, b). Signal ratios of adenine to purine (IA,734/IP,813) and hypoxanthine to purine (IHX,742/IP,813) (c, d), along with their corresponding RSD (d, f).
Figure 2c,2d displays the mean ratios of purine to adenine and hypoxanthine signals, respectively, derived from a total of 270 spectra taken from 27 spots on 3 different substrates. Both curves exhibit a steep slope in the low concentration ratio range, indicating poor sensitivity for predicting analyte concentrations when they are used as calibration curves. Conversely, they demonstrate a gentle slope in the higher-concentration ratio range, suggesting oversensitivity as calibration curves. Thus, to achieve the highest prediction accuracy, it is recommended to choose concentration ratios of purine/analyte between 1/3 and 3/1 to establish the calibration curve.
This choice is further supported by the RSDs of the measured concentration ratios of purine/adenine and purine/hypoxanthine, shown in Figure 2e,2f, respectively. The RSDs of the ratios reach a minimum of 10% when the concentration of IS matches that of the analyte. In contrast, the RSD for pure adenine is 30%, while for hypoxanthine it is 60%. These large RSDs are primarily attributed to the variation in the enhancement factors among the three randomly chosen substrates for these experiments, as the typical RSDs of our AgNP/AAO SERS substrates are 15–30% (see Figure S4).46
Figure S4a demonstrates that the use of purine as an IS effectively normalizes the spatial fluctuation of adenine signals on a SERS substrate. The normalized signal-map exhibits improved uniformity and the RSDs significantly reduce from 15–30 to 7.5%. The correlation coefficient between I734 from a pure adenine droplet and that from its neighboring mixture droplet on the substrate exhibits a moderate correlation (0.46–0.7), indicating that the use of an external standard on a neighboring site only moderately normalizes the point-to-point signal fluctuation on the SERS substrate (Table S1). In contrast, the correlation coefficients between I734 and I813 of the mixture are close to 1 (0.94–0.97), confirming the effectiveness of purine as an effective IS.
Both adenine and purine signals collected from substrates exposed to ambient air for varying durations exhibit a decrease over time, reaching asymptotic values after 2 days of exposure (Figure S4a,b). This decrease suggests a reaction between air and the AgNP surfaces of the substrate, leading to degradation of the SERS enhancement factor for both molecules. The similar degradation enables the signal ratios to remain constant with smaller RSDs (Figure S4c). These findings provide further confirmation of purine’s effectiveness as an IS for quantifying analytes using substrates exposed to air for up to 2 days. Since air exposure is unavoidable in practical SERS applications, the efficacy of purine as an IS is further validated.
Calibration curves were established for purine across 5 orders of concentration by maintaining the IS/analyte ratio between 3/10 and 3, as shown in Figure 3b,3e. For comparison, the signals of pure adenine and hypoxanthine were also recorded, as shown in Figure 3a,3d. (The data were acquired from 9 separate spots on 3 different substrates.) All curves exhibit a monotonically increasing trend, ensuring their suitability for predicting the concentrations of unknown analytes. However, the curves derived from the signals exhibit large fluctuations, indicating their inaccuracy for the concentration predictions. In contrast, the calibration curves derived from the analyte/IS signal ratios demonstrate reduced fluctuations and good linearity, enabling more precise and accurate predictions.
Figure 3.
Calibration curves for adenine and hypoxanthine derived without (a, d) and with (b, e) purine IS and their corresponding RSDs (c, f). The calibration curves for pure adenine (IA,734) and hypoxanthine (d) (IHX,742) at various concentrations and shown in (a) and (d), respectively. The curves in (b, e) display, respectively, the signal ratios of adenine to purine (IA,734/IP,813) and hypoxanthine to purine (IHX,742/IP,813) at various IS concentrations, while maintaining IS/analyte ratio between 3/10 and 3.
Figure 3c,3f displays the RSDs for adenine and hypoxanthine, respectively, demonstrating the improved accuracy achieved when purine is used as an IS to establish the calibration curves. The data clearly show the expected improvement in accuracy when purine is employed as an IS for establishing the calibration curves. Notably, the RSDs decrease with increasing analyte concentrations, consistent with the statistical nature of the fluctuation. Furthermore, the slope of the calibration curves for adenine with IS (Figure 3b) changes abruptly as the concentration of purine increases from 10–6 to 10–5 M, suggesting a switching of the adsorption competition between adenine and purine in this concentration range on the AgNP surface. In contrast, no such dramatic slope change is observed for hypoxanthine (Figure 3e). It appears that, above a concentration of 10–5 M, the adsorption isotherm for adenine exhibits a sharp increase, while those for purine and hypoxanthine remain similar. This suggests the occurrence of either multilayer adsorption (Brunauer–Emmett–Teller-type adsorption)47 or a change in adsorption geometry on the AgNP surface at higher adenine concentrations.48 Further experimental and theoretical studies are needed to clarify this long-standing controversy about the adsorption behavior of adenine on silver surfaces.
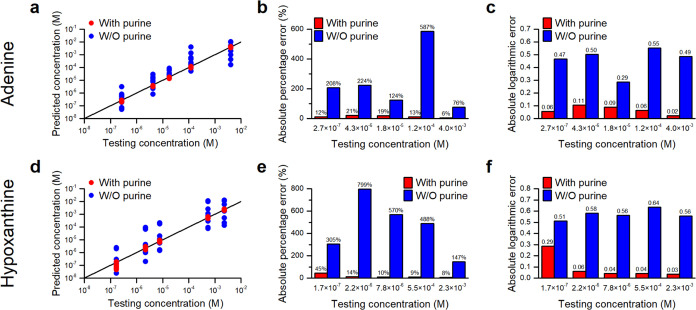
Figure 4a,d depicts the prediction capabilities of calibration curves derived with and without purine IS for various randomly selected concentrations of adenine and hypoxanthine, respectively. The data clearly exhibit improved accuracy and reduced variance of predictions derived from calibration curves with IS. To compare the prediction capabilities quantitatively, Figure 4b,4e shows the mean absolute percentage errors of the predictions. Overall, a more than 10-fold improvement in prediction accuracy is achieved with the use of purine IS.
Figure 4.
Prediction capability of the calibration curves with and without purine IS for various concentrations of (a–c) adenine and (d–f) hypoxanthine. (a, d) Predicted concentrations vs tested concentrations, (b, e) average absolute percentage errors, and (c, f) average absolute logarithmic errors.
Although the mean absolute percentage errors provide a general assessment of the accuracy of the predictions, they fail to reflect the seriousness of the error made when a predicted concentration is much smaller than the test concentration. It saturates at 100% when the former approaches zero and makes a limited contribution to the average percentage errors in the prediction accuracy. Therefore, we propose employing an absolute logarithmic error, defined as
to reflect the accuracy of a prediction. This error reveals how far off a prediction is on a logarithmic scale and is useful for assessing the accuracy of a prediction in terms of orders of magnitude. Figure 4c,4f shows the means of the absolute logarithmic errors for the predictions for adenine and hypoxanthine. Without purine IS, the means are close to or higher than 1/2, indicating that the predictions are either ∼3 times higher or ∼1/3 times smaller than the actual prediction on average. In contrast, with purine IS, the means are smaller than 0.11, except in one case. In other words, the predictions are within 0.77–1.3 times the testing concentrations. Notably, half of the predictions are within 0.91–1.1 times. The significant reduction in absolute logarithmic errors confirms the effectiveness of using purine as an IS for accurately predicting concentrations of both adenine and hypoxanthine.
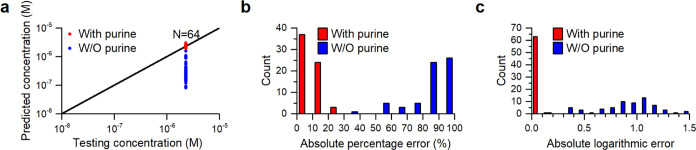
Figure 5 demonstrates the long-term effectiveness of using purine IS for predicting adenine concentration based on its SERS signals obtained from 64 substrates, which were fabricated in eight separate batches over six months after the derivation of calibration curves using data collected from three substrates. The results confirm the calibration’s validity even after 6 months. In contrast, the predicted concentrations without purine IS exhibit not only a broad distribution but also a shift toward smaller values, indicating a systematic reduction in the enhancement factor of substrates fabricated after the 3-month period. The shift may originate from subtle changes in the sizes and interparticle distances of the nanoparticles on different substrates.
Figure 5.
Prediction capability of adenine calibration curves with or without purine IS on different batches of substrates fabricated over 6 months. (a) Scatter plot of predicted concentrations vs tested concentrations, (b) histogram of absolute percentage errors, and (c) histogram of absolute logarithmic errors.
Figure 5b,5c shows histograms of the absolute percentage and the logarithmic errors in the predictions. When using purine IS, the absolute percentage errors are mostly below 30%, with over half below 10%, and the absolute logarithmic errors are primarily between 0 and 0.1, with 2 rare exceptions between 0.1 and 0.2. Without IS, the majority of the prediction errors exceed 50%, with 50 out of 64 predictions deviating by over 80%, and the absolute logarithmic errors range from 0.1 to 1.5. Essentially, the use of purine IS significantly improves prediction precision and accuracy from an order of magnitude estimates to a few tens of percent. These findings also demonstrate the continued validity of the prediction curves derived from purine IS, even after 6 months.
The effectiveness of purine as a SERS IS for quantifying purine derivative metabolites released by bacteria was tested by comparing the concentrations of adenine and hypoxanthine measured in the supernatants of S. aureus and E. coli with those measured by LC-MS, which has been demonstrated to provide reliable measurements previously.12Figure 6a,6c shows that SERS measurements with purine IS closely matched LC-MS measurements, whereas measurements without IS were significantly different. Figure 6b,d demonstrates the accuracy of SERS measurements with IS, which were within a few tens of percent, as opposed to the lesser accuracy of SERS measurements without IS, deviating by almost an order of magnitude in most cases and two orders in extreme cases. These discrepancies can be attributed to a systematic long-term reduction in the SERS-enhancing power of substrates fabricated after the calibration curves were acquired from earlier substrates. It should be noted that concentrations measured by SERS with IS were slightly higher than those by LC-MS, except in one case, likely due to contributions from other adenine derivatives such as AMP in the bacterial supernatants. However, this systematic error is manageable as AMP concentrations in bacterial supernatants have been shown to be an order of magnitude lower than adenine and hypoxanthine.11,12 Overall, these results indicate that employing purine IS enables accurate quantification of adenine and hypoxanthine in bacterial supernatants using SERS substrates fabricated over a long period, despite their fluctuating enhancement factors.
Figure 6.
Quantification of adenine and hypoxanthine released, respectively, by S. aureus and E. coli using SERS with and without purine IS, and comparison with LC-MS measurements. (a, c) Measured concentrations of adenine and hypoxanthine, respectively. (b, d) Ratios of SERS-measured concentrations to LC-MS measurements.
The data above clearly demonstrate that purine effectively serves as an IS to enhance the precision and accuracy of the quantification of adenine and hypoxanthine in water and bacterial supernatants. Purine’s effectiveness can be attributed to its minimal interaction with the analytes, avoiding complications such as peak shifting and additional peak formation in the spectra. Additionally, the signals of purine and the analytes are well separated, enabling a clear distinction between their signals. Furthermore, purine’s adsorption onto the AgNP/AAO substrates preserves the substrate’s surface chemistry to the extent that the adsorption behavior of the analytes on individual AgNP surfaces and in the hot spots between neighboring AgNPs remains similar in the analyte-IS mixture. Essentially, the adsorption of IS and analyte molecules onto the substrate’s surface can be seen as competing events for available sites, with both having a similar probability of adsorbing onto various nanostructures’ surfaces that continue to evolve when exposed to water with analytes or ambient air.
Despite the significant variation in their enhancement factors, the surface chemistry of different batches of substrates fabricated over a half-year period exhibited such similarity that the IS-normalized calibration curves derived from the first batch of substrates remained valid. The validity implies the robustness of the fabrication process for the AgNP/AAO SERS substrates.
Using purine IS in practical bacterial applications offers several advantages. First, it minimizes fluctuations, reducing errors and the need for additional measurements. Second, purine IS can effectively normalize the declining SERS signals from a substrate that was exposed to ambient air for up to 2 days, ensuring consistent results, which is especially valuable for multiple measurements on large-scale substrates with varying enhancements in such conditions. Additionally, the 6-month durability of the calibration curves across substrate batches reduces the need for frequent recalibration, making it beneficial for repetitive applications on numerous substrates. While adding the step of mixing the IS molecule and SERS measurements may slightly extend the workflow, it typically requires less than 10 additional minutes when the analyte’s concentration order of magnitude is unknown, and this time can be further reduced to a few minutes if the concentration order is already known. Since purine is not naturally occurring, utilizing it as an IS holds promise for accurate, real-time, in situ quantification of adenine and hypoxanthine released from bacteria under various stress conditions, including starvation and antibiotic exposure.
In principle, purine can potentially function as an IS on other solid SERS substrates for quantifying purine derivatives due to their chemical similarity. However, the validity of using purine as an IS depends on the surface chemistry of the nanostructures on the substrates. This factor determines whether a calibration curve obtained from one batch of substrates can accurately quantify purine derivative molecules by using a different batch of substrates fabricated later. The suitability of purine as an IS on colloidal SERS substrates is even more uncertain, as the presence of purine molecules in the analyte solution may affect the formation of hot spots between pairs of colloids, leading to an increased signal fluctuation. Further studies are required to clarify the applicability and validity of purine as an IS on various SERS substrates.
Conclusions
Our study unequivocally demonstrates the effectiveness and convenience of purine as an IS for the accurate SERS quantification of purine derivatives. Purine’s distinctive ring-breathing-mode peak, minimal interaction with adenine/hypoxanthine, and ease of accessibility contribute to its suitability as an IS. By employing purine as an IS, we significantly improved measurement accuracy across a wide concentration range of analytes and clearly showed the robustness of calibration curves for up to six months due to the consistent surface chemistry of our AgNP/AAO SERS substrates. Notably, purine remains a reliable IS even under practical conditions, including prolonged exposure of the substrate to ambient air, simplifying operational procedures, and enhancing the applicability of the quantitative SERS measurement process.
Furthermore, we successfully utilize purine as an IS to quantify adenine and hypoxanthine in the supernatants of S. aureus and E. coli, respectively, obtaining measurements that closely align with those obtained by LC-MS. The fact that “purine itself has never been found in nature” renders it a versatile IS applicable to a wide range of biological samples. This significant finding represents a notable advancement in quantitative SERS measurements of extracellular metabolites, providing a foundation for the in situ SERS quantification of metabolites released by microorganisms under various stress conditions. This development holds great potential for exploring the metabolic dynamics and responses of microorganisms in complex environments.
Acknowledgments
This research was supported by the National Science and Technology Council (MOST 111-2123-M-001-005) and Academia Sinica in Taiwan. The authors thank the assistance from the Small Molecule Metabolomics Core Facility sponsored by the Institute of Plant and Microbial Biology, Academia Sinica for LC-MS analysis.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.3c03259.
Brief introduction of SERS-AST workflow; scanning electron microscopy (SEM) image of Ag/AAO SERS substrate, SERS spectra of the pure purines; maps of adenine signals and signal ratios with and without purine; flowcharts of bacterial supernatant preparation and concentration measurement protocol via SERS with purine; calibration curves by LC-MS; and chromatographic peak areas of the purines and the supernatants by LC-MS (PDF)
Author Contributions
H.-W.C. and Y.-L.W. designed the experiments. H.-W.C. and H.-M.T. performed the experiments. H.-W.C. analyzed the data and prepared the manuscript. Y.-L.W. proposed the original idea, guided the experiments, and revised the manuscript. All authors have approved the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Yan M.; Li H.; Li M.; Cao X.; She Y.; Chen Z. Advances in Surface-Enhanced Raman Scattering-Based Aptasensors for Food Safety Detection. J. Agric. Food Chem. 2021, 69 (47), 14049–14064. 10.1021/acs.jafc.1c05274. [DOI] [PubMed] [Google Scholar]
- Xu G. D.; Song P.; Xia L. X. Examples in the detection of heavy metal ions based on surface-enhanced Raman scattering spectroscopy. Nanophotonics 2021, 10 (18), 4419–4445. 10.1515/nanoph-2021-0363. [DOI] [Google Scholar]
- Yaraki M. T.; Tukova A.; Wang Y. Emerging SERS biosensors for the analysis of cells and extracellular vesicles. Nanoscale 2022, 14 (41), 15242–15268. 10.1039/D2NR03005E. [DOI] [PubMed] [Google Scholar]
- Tahir M. A.; Dina N. E.; Cheng H.; Valev V. K.; Zhang L. Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis. Nanoscale 2021, 13 (27), 11593–11634. 10.1039/D1NR00708D. [DOI] [PubMed] [Google Scholar]
- Dina N. E.; Tahir M. A.; Bajwa S. Z.; Amin I.; Valev V. K.; Zhang L. SERS-based antibiotic susceptibility testing: Towards point-of-care clinical diagnosis. Biosens. Bioelectron. 2023, 219, 114843 10.1016/j.bios.2022.114843. [DOI] [PubMed] [Google Scholar]
- Samek O.; Bernatová S.; Dohnal F. The potential of SERS as an AST methodology in clinical settings. Nanophotonics 2021, 10 (10), 2537–2561. 10.1515/nanoph-2021-0095. [DOI] [Google Scholar]
- Liu C. Y.; Han Y. Y.; Shih P. H.; Lian W. N.; Wang H. H.; Lin C. H.; Hsueh P. R.; Wang J. K.; Wang Y. L. Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Sci. Rep. 2016, 6, 23375 10.1038/srep23375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. Y.; Lin Y. C.; Cheng W. C.; Lin Y. T.; Teng L. J.; Wang J. K.; Wang Y. L. Rapid antibiotic susceptibility testing of bacteria from patients’ blood via assaying bacterial metabolic response with surface-enhanced Raman spectroscopy. Sci. Rep. 2020, 10 (1), 12538 10.1038/s41598-020-68855-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-Y.; Wang J.-T.; Cheng W.-C.; Chen K.-L.; Chi Y.; Teng L.-J.; Wang J.-K.; Wang Y.-L. SERS-based rapid susceptibility testing of commonly administered antibiotics on clinically important bacteria species directly from blood culture of bacteremia patients. World J. Microbiol. Biotechnol. 2023, 39 (10), 282 10.1007/s11274-023-03717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J. L.; Ikuta K. S.; Sharara F.; Swetschinski L.; Robles Aguilar G.; Gray A.; Han C.; Bisignano C.; Rao P.; Wool E. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399 (10325), 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premasiri W. R.; Lee J. C.; Sauer-Budge A.; Theberge R.; Costello C. E.; Ziegler L. D. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal. Bioanal. Chem. 2016, 408 (17), 4631–4647. 10.1007/s00216-016-9540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W. Y.; Cheng H. W.; Chen Z. X.; Wang H. H.; Lai M. Y.; Wang J. K.; Wang Y. L. Quantification of biomolecules responsible for biomarkers in the surface-enhanced Raman spectra of bacteria using liquid chromatography-mass spectrometry. Phys. Chem. Chem. Phys. 2018, 20 (12), 8032–8041. 10.1039/C7CP07103E. [DOI] [PubMed] [Google Scholar]
- Langer J.; Jimenez de Aberasturi D.; Aizpurua J.; Alvarez-Puebla R. A.; Auguie B.; Baumberg J. J.; Bazan G. C.; Bell S. E. J.; Boisen A.; Brolo A. G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14 (1), 28–117. 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrimsek A. B.; Chiang N.; Mattei M.; Zaleski S.; McAnally M. O.; Chapman C. T.; Henry A. I.; Schatz G. C.; Van Duyne R. P. Single-Molecule Chemistry with Surface- and Tip-Enhanced Raman Spectroscopy. Chem. Rev. 2017, 117 (11), 7583–7613. 10.1021/acs.chemrev.6b00552. [DOI] [PubMed] [Google Scholar]
- Bell S. E. J.; Sirimuthu N. M. Quantitative surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2008, 37 (5), 1012–1024. 10.1039/b705965p. [DOI] [PubMed] [Google Scholar]
- Wang H. H.; Liu C. Y.; Wu S. B.; Liu N. W.; Peng C. Y.; Chan T. H.; Hsu C. F.; Wang J. K.; Wang Y. L. Highly Raman-enhancing substrates based on silver nanoparticle arrays with tunable sub-10 nm gaps. Adv. Mater. 2006, 18 (4), 491–495. 10.1002/adma.200501875. [DOI] [Google Scholar]
- Zhang Q.; Li J.; Tang P.; Lu X.; Tian J.; Zhong L. Dynamic Imaging of Transferrin Receptor Molecules on Single Live Cell with Bridge Gaps-Enhanced Raman Tags. Nanomaterials 2019, 9 (10), 1373 10.3390/nano9101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman S. J.; Darweesh A. A.; Furr M.; Magee M.; Argyropoulos C.; Herzog J. B. Tunable SERS Enhancement via Sub-nanometer Gap Metasurfaces. ACS Appl. Mater. Interfaces 2022, 14 (13), 15541–15548. 10.1021/acsami.2c01335. [DOI] [PubMed] [Google Scholar]
- Le Ru E. C.; Etchegoin P. G.; Meyer M. Enhancement factor distribution around a single surface-enhanced Raman scattering hot spot and its relation to single molecule detection. J. Chem. Phys. 2006, 125 (20), 204701 10.1063/1.2390694. [DOI] [PubMed] [Google Scholar]
- Fang Y.; Seong N. H.; Dlott D. D. Measurement of the distribution of site enhancements in surface-enhanced Raman scattering. Science 2008, 321 (5887), 388–392. 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- Grys D. B.; Chikkaraddy R.; Kamp M.; Scherman O. A.; Baumberg J. J.; Nijs B. Eliminating irreproducibility in SERS substrates. J. Raman Spectrosc. 2021, 52 (2), 412–419. 10.1002/jrs.6008. [DOI] [Google Scholar]
- Chen W. L.; Lo C. Y.; Huang Y. C.; Wang Y. C.; Chen W. H.; Lin K. J.; Chang Y. M. Toward quantitative SERS detection in low analyte concentration by investigating the immersion volume and time of SERS substrate in analyte solution. J. Raman Spectrosc. 2022, 53 (1), 33–39. 10.1002/jrs.6250. [DOI] [Google Scholar]
- Kahl M.; Voges E.; Kostrewa S.; Viets C.; Hill W. Periodically structured metallic substrates for SERS. Sens. Actuators, B 1998, 51 (1–3), 285–291. 10.1016/S0925-4005(98)00219-6. [DOI] [Google Scholar]
- Fu Q.; Zhan Z.; Dou J.; Zheng X.; Xu R.; Wu M.; Lei Y. Highly Reproducible and Sensitive SERS Substrates with Ag Inter-Nanoparticle Gaps of 5 nm Fabricated by Ultrathin Aluminum Mask Technique. ACS Appl. Mater. Interfaces 2015, 7 (24), 13322–13328. 10.1021/acsami.5b01524. [DOI] [PubMed] [Google Scholar]
- Garg A.; Nam W.; Zhou W. Reusable Surface-Enhanced Raman Spectroscopy Membranes and Textiles via Template-Assisted Self-Assembly and Micro/Nanoimprinting. ACS Appl. Mater. Interfaces 2020, 12 (50), 56290–56299. 10.1021/acsami.0c16351. [DOI] [PubMed] [Google Scholar]
- Song J.; Nam W.; Zhou W. Scalable High-Performance Nanolaminated SERS Substrates Based on Multistack Vertically Oriented Plasmonic Nanogaps. Adv. Mater. Technol. 2019, 4 (5), 1800689 10.1002/admt.201800689. [DOI] [Google Scholar]
- Mejia E.; Song J.; Zhao Y.; Qian Y.; Xiao C.; Lezec H. J.; Agrawal A.; Zhou W. Scalable two-tier protruding micro-/nano-optoelectrode arrays with hybrid optical-electrical modalities by hierarchical modular design. Nanoscale 2022, 14 (41), 15373–15383. 10.1039/D2NR03820J. [DOI] [PubMed] [Google Scholar]
- Nam W.; Song J.; Ali Safiabadi Tali S.; Lezec H. J.; Agrawal A.; Zhou W. Au/SiO2-Nanolaminated Plasmonic Nanoantennas as Refractive-Index-Insensitive and Transparent Surface-Enhanced Raman Spectroscopy Substrates. ACS Appl. Nano Mater. 2021, 4 (3), 3175–3184. 10.1021/acsanm.1c00389. [DOI] [Google Scholar]
- Simoncelli S.; Roller E. M.; Urban P.; Schreiber R.; Turberfield A. J.; Liedl T.; Lohmuller T. Quantitative Single-Molecule Surface-Enhanced Raman Scattering by Optothermal Tuning of DNA Origami-Assembled Plasmonic Nanoantennas. ACS Nano 2016, 10 (11), 9809–9815. 10.1021/acsnano.6b05276. [DOI] [PubMed] [Google Scholar]
- Li L.; Wang Z.; Lu Y.; Zhu K.; Zong S.; Cui Y. DNA-assisted synthesis of Ortho-NanoDimer with sub-nanoscale controllable gap for SERS application. Biosens. Bioelectron. 2021, 172, 112769 10.1016/j.bios.2020.112769. [DOI] [PubMed] [Google Scholar]
- Zhang D.; Xie Y.; Deb S. K.; Davison V. J.; Ben-Amotz D. Isotope edited internal standard method for quantitative surface-enhanced Raman spectroscopy. Anal. Chem. 2005, 77 (11), 3563–3569. 10.1021/ac050338h. [DOI] [PubMed] [Google Scholar]
- Tian H. H.; Zhang N.; Zhang J.; Tong L. M. Exploring quantification in a mixture using graphene-based surface-enhanced Raman spectroscopy. Appl. Mater. Today 2019, 15, 288–293. 10.1016/j.apmt.2019.01.008. [DOI] [Google Scholar]
- Lin S.; Hasi W.; Lin X.; Han S.; Xiang T.; Liang S.; Wang L. Lab-On-Capillary Platform for On-Site Quantitative SERS Analysis of Surface Contaminants Based on Au@4-MBA@Ag Core-Shell Nanorods. ACS Sens. 2020, 5 (5), 1465–1473. 10.1021/acssensors.0c00398. [DOI] [PubMed] [Google Scholar]
- Nam W.; Zhao Y.; Song J.; Ali Safiabadi Tali S.; Kang S.; Zhu W.; Lezec H. J.; Agrawal A.; Vikesland P. J.; Zhou W. Plasmonic Electronic Raman Scattering as Internal Standard for Spatial and Temporal Calibration in Quantitative Surface-Enhanced Raman Spectroscopy. J. Phys. Chem. Lett. 2020, 11 (22), 9543–9551. 10.1021/acs.jpclett.0c03056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam W.; Ren X.; Kim I.; Strobl J.; Agah M.; Zhou W. Plasmonically Calibrated Label-Free Surface-Enhanced Raman Spectroscopy for Improved Multivariate Analysis of Living Cells in Cancer Subtyping and Drug Testing. Anal. Chem. 2021, 93 (10), 4601–4610. 10.1021/acs.analchem.0c05206. [DOI] [PubMed] [Google Scholar]
- Lorén A.; Engelbrektsson J.; Eliasson C.; Josefson M.; Abrahamsson J.; Johansson M.; Abrahamsson K. Internal standard in surface-enhanced Raman spectroscopy. Anal. Chem. 2004, 76 (24), 7391–7395. 10.1021/ac0491298. [DOI] [PubMed] [Google Scholar]
- Anderson W. J.; Nowinska K.; Hutter T.; Mahajan S.; Fischlechner M. Tuning plasmons layer-by-layer for quantitative colloidal sensing with surface-enhanced Raman spectroscopy. Nanoscale 2018, 10 (15), 7138–7146. 10.1039/C7NR06656B. [DOI] [PubMed] [Google Scholar]
- Li G. Q.; Hao Q.; Li M. Z.; Zhao X.; Song W. Z.; Fan X. C.; Qiu T. Quantitative SERS Analysis by Employing Janus Nanoparticles with Internal Standards. Adv. Mater. Interfaces 2023, 10 (7), 2202127 10.1002/admi.202202127. [DOI] [Google Scholar]
- Liebeke M.; Dorries K.; Zuhlke D.; Bernhardt J.; Fuchs S.; Pane-Farre J.; Engelmann S.; Volker U.; Bode R.; Dandekar T.; et al. A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol. Biosyst. 2011, 7 (4), 1241–1253. 10.1039/c0mb00315h. [DOI] [PubMed] [Google Scholar]
- Bennett B. D.; Kimball E. H.; Gao M.; Osterhout R.; Van Dien S. J.; Rabinowitz J. D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5 (8), 593–599. 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmane R. S.Ring-Expanded (‘Fat‘) Purines and their Nucleoside/Nucleotide Analogues as Broad-Spectrum Therapeutics. In Progress in Heterocyclic Chemistry; Gribble G. W.; Joule J. A., Eds.; Elsevier, 2009; Vol. 21, pp 35–68. [Google Scholar]
- Baba T.; Ara T.; Hasegawa M.; Takai Y.; Okumura Y.; Baba M.; Datsenko K. A.; Tomita M.; Wanner B. L.; Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006, 2 (1), 0008 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morháč M. An algorithm for determination of peak regions and baseline elimination in spectroscopic data. Nucl. Instrum. Methods Phys. Res., Sect. A 2009, 600 (2), 478–487. 10.1016/j.nima.2008.11.132. [DOI] [Google Scholar]
- Mosier-Boss P. A. Review on SERS of Bacteria. Biosensors 2017, 7 (4), 51 10.3390/bios7040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Li W.; Sun M.; Wang H. Application of SERS in the Detection of Fungi, Bacteria and Viruses. Nanomaterials 2022, 12 (20), 3572 10.3390/nano12203572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvoynenko O.; Lo S. L.; Chen Y. J.; Chen G. W.; Tsai H. M.; Wang Y. L.; Wang J. K. Speciation Analysis of Cr(VI) and Cr(III) in Water with Surface-Enhanced Raman Spectroscopy. ACS Omega 2021, 6 (3), 2052–2059. 10.1021/acsomega.0c05020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunauer S.; Emmett P. H.; Teller E. Adsorption of Gases in Multimolecular Layers. J. Am. Chem. Soc. 1938, 60 (2), 309–319. 10.1021/ja01269a023. [DOI] [Google Scholar]
- Harroun S. G. The Controversial Orientation of Adenine on Gold and Silver. ChemPhysChem 2018, 19 (9), 1003–1015. 10.1002/cphc.201701223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.