Abstract
Two biosynthetic routes to the heme, chlorophyll, and phycobilin precursor, δ-aminolevulinic acid (ALA) are known: conversion of the intact five-carbon skeleton of glutamate, and ALA synthase-catalyzed condensation of glycine plus succinyl-coenzyme A. The existence and physiological roles of the two pathways in Cyanidium caldarium were assessed in vivo by determining the relative abilities of [2-14C]glycine and [1-14C]glutamate to label protoheme and heme a. Glutamate was incorporated to a much greater extent than glycine into both protoheme and heme a, even in cells that were unable to form chlorophyll and phycobilins. The small incorporation of glycine could be accounted for by transfer of label to intracellular glutamate pools, as determined from amino acid analysis. It thus appears that C. caldarium makes all tetrapyrroles, including mitochondrial hemes, solely from glutamate, and there is no contribution by ALA synthase in this organism.
Full text
PDF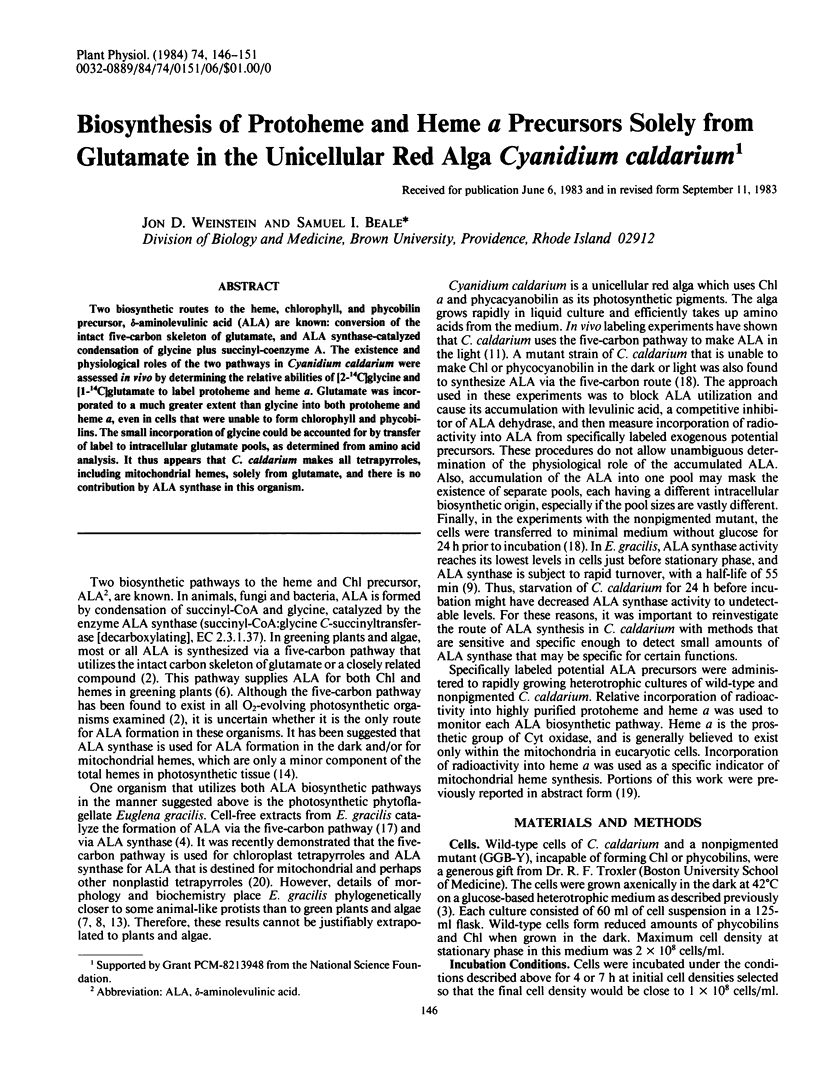


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beale S. I., Chen N. C. N-Methyl Mesoporphyrin IX Inhibits Phycocyanin, but Not Chlorophyll Synthesis in Cyanidium caldarium. Plant Physiol. 1983 Feb;71(2):263–268. doi: 10.1104/pp.71.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale S. I., Foley T., Dzelzkalns V. delta-Aminolevulinic acid synthase from Euglena gracilis. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1666–1669. doi: 10.1073/pnas.78.3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale S. I. Studies on the Biosynthesis and Metabolism of delta-Aminolevulinic Acid in Chlorella. Plant Physiol. 1971 Sep;48(3):316–319. doi: 10.1104/pp.48.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson J. R., Hare P. E. O-phthalaldehyde: fluorogenic detection of primary amines in the picomole range. Comparison with fluorescamine and ninhydrin. Proc Natl Acad Sci U S A. 1975 Feb;72(2):619–622. doi: 10.1073/pnas.72.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelfranco P. A., Jones O. T. Protoheme turnover and chlorophyll synthesis in greening barley tissue. Plant Physiol. 1975 Mar;55(3):485–490. doi: 10.1104/pp.55.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Eukaryote kingdoms: seven or nine? Biosystems. 1981;14(3-4):461–481. doi: 10.1016/0303-2647(81)90050-2. [DOI] [PubMed] [Google Scholar]
- Chang S. H., Hecker L. I., Brum C. K., Schnabel J. J., Heckman J. E., Silberklang M., RajBhandary U. L., Barnett W. E. The nucleotide sequence of Euglena cytoplasmic phenylalanine transfer RNA. Evidence for possible classifications of Euglena among the animal rather than the plant kingdom. Nucleic Acids Res. 1981 Jul 10;9(13):3199–3204. doi: 10.1093/nar/9.13.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley T., Dzelzkalns V., Beale S. I. delta-Aminolevulinic Acid Synthase of Euglena gracilis: Regulation of Activity. Plant Physiol. 1982 Jul;70(1):219–226. doi: 10.1104/pp.70.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke U. Tritosol: a new scintillation cocktail based on Triton X-100. Anal Biochem. 1975 Feb;63(2):555–558. doi: 10.1016/0003-2697(75)90379-6. [DOI] [PubMed] [Google Scholar]
- Jurgenson J. E., Beale S. I., Troxler R. F. Biosynthesis of delta-aminolevulinic acid in the unicellular rhodophyte, cyanidium caldarium. Biochem Biophys Res Commun. 1976 Mar 8;69(1):149–157. doi: 10.1016/s0006-291x(76)80285-9. [DOI] [PubMed] [Google Scholar]
- Klein O., Senger H. Two Biosynthetic Pathways to delta-Aminolevulinic Acid in a Pigment Mutant of the Green Alga, Scenedesmus obliquus. Plant Physiol. 1978 Jul;62(1):10–13. doi: 10.1104/pp.62.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S. Nucleotide sequence of cytoplasmic 5S ribosomal RNA from Euglena gracilis. J Mol Evol. 1982;18(5):293–296. doi: 10.1007/BF01733894. [DOI] [PubMed] [Google Scholar]
- Oh-hama T., Seto H., Otake N., Miyachi S. 13c-NMR evidence for the pathway of chlorophyll biosynthesis in green algae. Biochem Biophys Res Commun. 1982 Mar 30;105(2):647–652. doi: 10.1016/0006-291x(82)91483-8. [DOI] [PubMed] [Google Scholar]
- Porra R. J., Klein O., Wright P. E. The proof by 13C-NMR spectroscopy of the predominance of the C5 pathway over the Shemin pathway in chlorophyll biosynthesis in higher plants and of the formation of the methyl ester group of chlorophyll from glycine. Eur J Biochem. 1983 Feb 15;130(3):509–516. doi: 10.1111/j.1432-1033.1983.tb07179.x. [DOI] [PubMed] [Google Scholar]
- Troxler R. F., Offner G. D. delta-Aminolevulinic acid synthesis in a Cyanidium caldarium mutant unable to make chlorophyll a and phycobiliproteins. Arch Biochem Biophys. 1979 Jun;195(1):53–65. doi: 10.1016/0003-9861(79)90326-6. [DOI] [PubMed] [Google Scholar]
- Weinstein J. D., Beale S. I. Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J Biol Chem. 1983 Jun 10;258(11):6799–6807. [PubMed] [Google Scholar]


