Extended Data Figure 11. Old nmrHAS2 mice differ from age matched CreER controls in their gut microbiome composition.
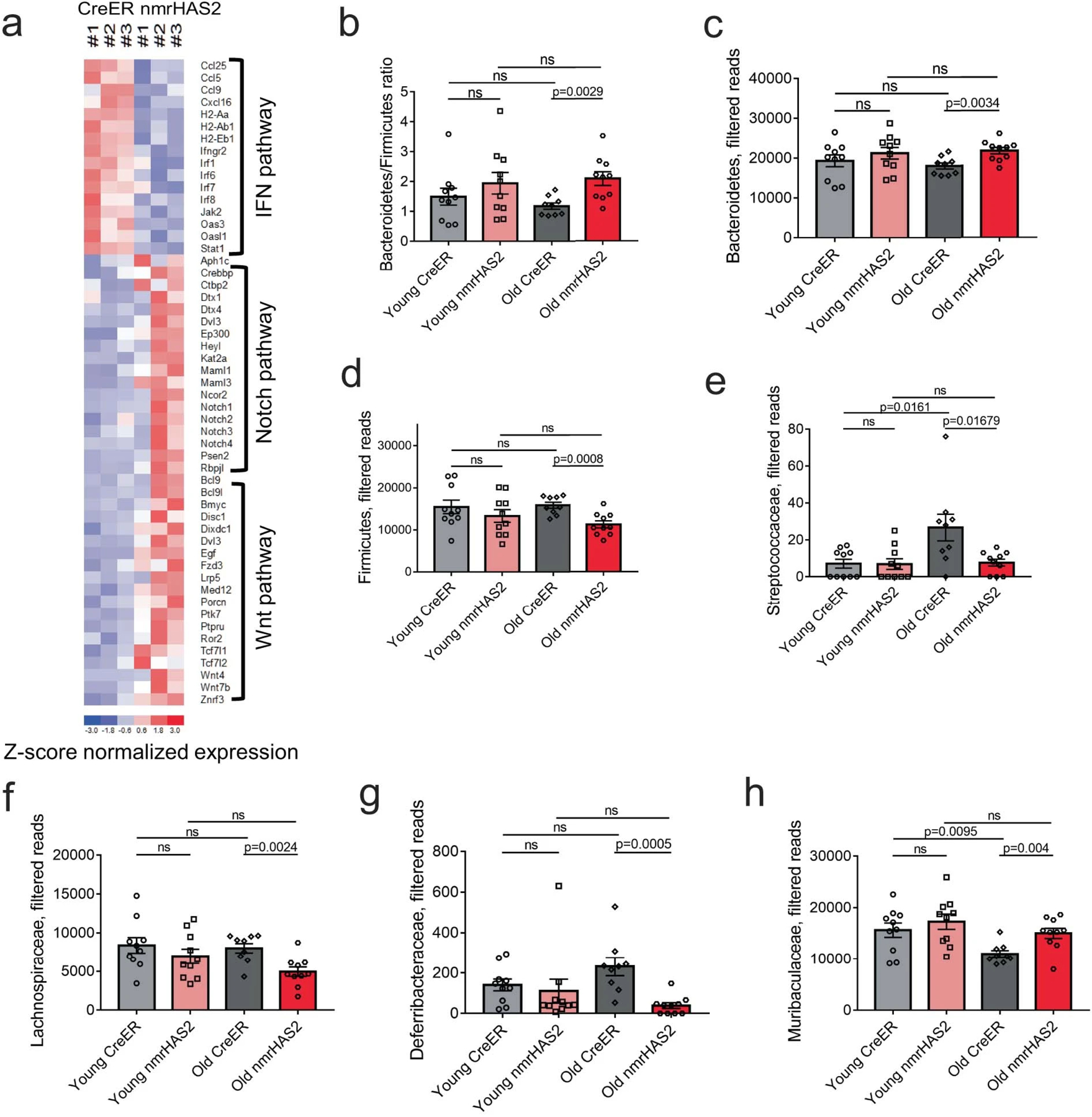
a. Heatmap of genes involve in IFN, WNT, and Notch pathways.
b.16s rRNA sequencing shows old nmrHAS2 mice (n=9) have a higher B/F ratio compared to age-matched controls (n=10). Pooled females and males.
c. 16s rRNA sequencing shows that at the phylum level old nmrHAS2 mice (n=9) have more abundant Bacteroidetes and less abundant Firmicutes compared to age-matched controls (n=10). 7- and 24-month-old mice were used. Pooled females and males.
d. 16s rRNA sequencing shows that at family level, old nmrHAS2 mice (n=9) have less abundant pro-inflammatory Streptococcaceae, Lachnospiraceae, and Deferribacteraceae compared to age-matched controls (n=10). 7- and 24-month-old mice were used. Pooled females and males.
e. 16s rRNA sequencing shows that at family level, old nmrHAS2 mice (n=9) have more Muribaculaceae compared to the age-matched controls (n=10). Pooled females and males.
b-e. p-values were calculated by two-tailed unpaired t-test (p-values are indicated in the graphs). Bars represent the means, error bars show standard errors, dots represent biological replicates. Adjustments were made for multiple comparisons.
