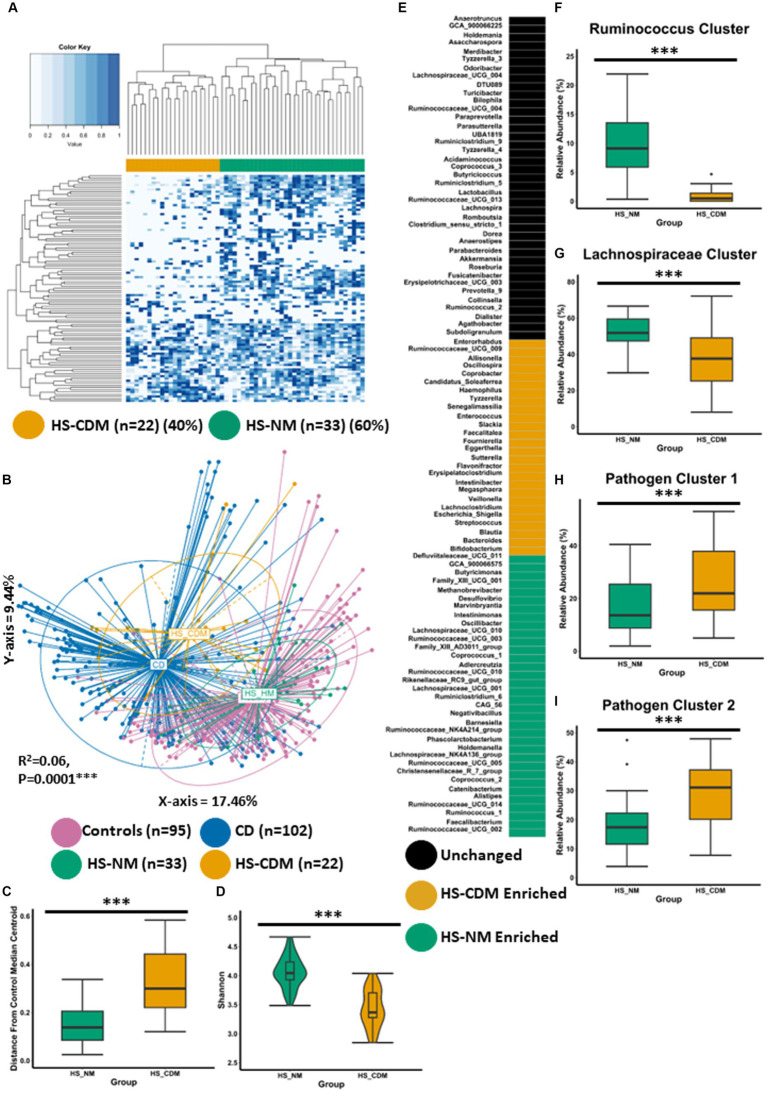
Figure 4.
Some patients with HS have a microbiota configuration similar to patients with CD. (A) Heatmap showing the genus profiles of all patients with HS. Through Ward.d2 clustering two genus level microbiome groups were identified labelled HS-NM (HS-Healthy-like microbiota; green) and HS-CDM (HS-Crohns disease-like microbiota; yellow). (B) Principal Component Analysis (PCoA) of β-diversity (Bray–Curtis dissimilarity) at the genus level (16S rRNA gene amplicon profiles). The p-value (0.0001) obtained using a PERMONOVA shows there is statistically significant microbiome separation between the groups even after controlling for the study effect and patient identifier as confounders. The eigen values are also reported which show the variation reported in the X-axis (17.46%) and Y-axis (9.44%) of the PCoA. (C) Using the PCoA coordinates from (A) the median control centroid was calculated and the distance of samples (HS-HM and HS-CDM) from those coordinates was subsequently determined and displayed here as a box plot. (D) Boxplot showing Shannon microbiome α-diversity between the groups. (E) ANCOMBC differential abundance analysis was sued to determine significantly differentially abundant genera between the groups. Each colour signifies whether that specific genera was unchanged (black), higher in HS-HM (green), higher HS-CDM (yellow). (F–I) Boxplots showing the relative abundance (%) of each CAG across the two groups of patients with HS. Wilcoxon test was used to determine significant differences between the groups. The annotations used for p-values are p < 0.05*; p < 0.01**; p < 0.001***. All displayed p-values are FDR corrected.