Figure 2.
Assembly of synXV by CRIMiRE
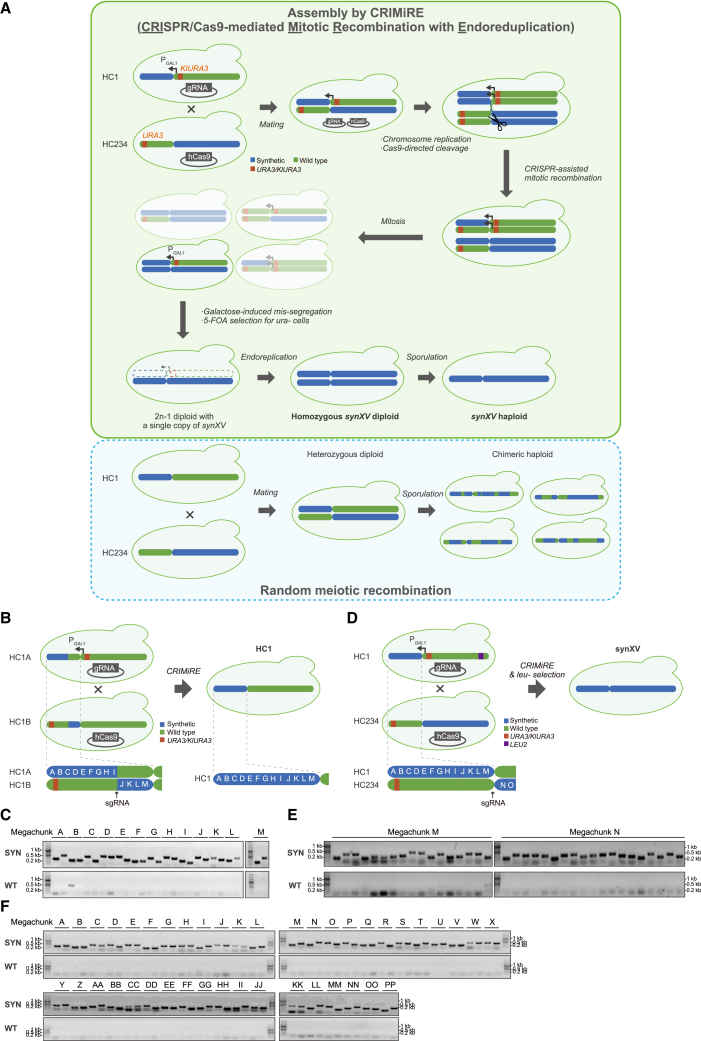
(A) CRIMiRE requires strategic modifications to the semisynthetic chromosomes as follows: (1) URA3 markers near the respective telomere and centromere within the wild-type segments to be removed and (2) a galactose-inducible promoter (PGAL1) upstream of the centromere within the wild-type segment to be removed. CRISPR-Cas9-mediated mitotic recombination commences upon mating of the two strains carrying plasmids for expressing Cas9 and a single guide RNA (sgRNA) separately. Assembled synXV coexists in a mixed population of heterozygous diploids. Galactose induction missegregates chromosomes with PGAL1 upstream of the centromere.13 Under 5-fluoroorotic acid (5-FOA) selection, hemizygous synXV diploids are the only Ura– cells within the missegregated population that can grow. Subsequent endoreduplication of the synXV chromosome in the 2n-1 strain results in a 2n homozygous synXV diploid, which is then sporulated to obtain a haploid synXV. In contrast, meiotic recombination would lead to a myriad of chimeric chromosomes, which would be difficult to screen to identify a strain with complete synXV.
(B) HC1A and HC1B were modified with a KlURA3-PGAL1-CEN15 cassette and URA3 from chunk A5 near the left terminus, respectively. These derived strains were consolidated into HC1 by CRIMiRE by cleaving between megachunks I and J. The resulting clones were screened by PCRTags (Figures S1F–S1H) to identify clones with homozygous HC1.
(C) Analysis of HC1 with synthetic and wild-type PCRTags (SYN and WT, respectively). A strain (Figures S1G–S1H, clone 7) showed only synthetic PCRTags in megachunks I and J. Further analysis with the first and last PCRTags of each megachunk showed the clone to be synthetic throughout HC1.
(D) HC1 was modified with a KlURA3-PGAL1-CEN15 cassette and a LEU2 from chunk NN4 near the right terminus. HC234 was modified with a URA3 from chunk A5 near the left terminus. These derived strains were consolidated into synXV by CRIMiRE by cleaving between megachunks M and N. The resulting clones were screened by auxotrophy and PCRTags (Figures S1J–S1N) to identify clones with homozygous synXV.
(E) A synXV candidate (Figures S1L–S1N, clone L6) was found to be synthetic and homozygous in megachunks M and N, which flanked the cleavage site.
(F) Two representative PCRTags from each megachunk showing that the synXV clone was synthetic and homozygous throughout the chromosome. A complete PCRTag analysis was performed (Figure S2).