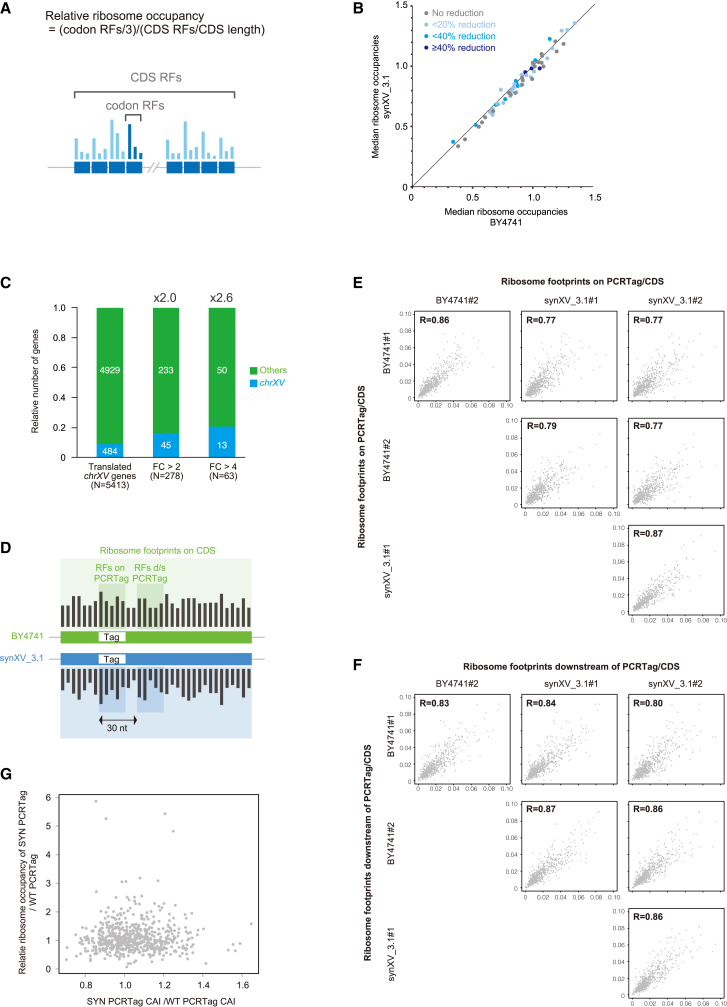
Figure 5.
Translatomic analysis of the synthetic yeast
(A) Schematic of relative ribosome occupancy (RRO). The blue boxes represent codons. The bars on the blue boxes represent the number of ribosome footprints (RFs) whose A site was mapped onto the codons.
(B) Scatterplot showing that loss of tRNA did not affect the translation elongation rate. Each dot represents the median value of the RROs for a codon sequence in all coding DNA sequences (CDSs) encoded on non-chrXV. The percentage reduction in the tRNA copy number in synXV relative to BY4741 was calculated for every corresponding codon sequence and is represented in different colors.
(C) Differentially translated genes were enriched in the synthetic chromosome in synXV_3.1. Translated genes were defined as genes with reads per kilobase per million mapped (RPKM) > 1 in BY4741 and synXV_3.1. FC, fold change.
(D) Schematic of the 30-nt regions on and downstream of each PCRTag analyzed in (E) and (F).
(E and F) Scatterplots showing a lower correlation of the translation elongation rate on PCRTags between BY4741 and synXV_3.1 (E) but not on 30-nt regions downstream of the PCRTags (F). RFs on PCRTags (or 30 nt downstream)/CDSs were defined as the number of RFs mapped onto PCRTag regions (or 30 nt downstream of the PCRTags) divided by those onto the corresponding CDSs. Sample numbers are indicated after the strain names (biological duplicates).
(G) Scatterplot showing no correlation between codon adaptation index (CAI) changes on the PCRTags and translation elongation changes on the PCRTags. CAIs of all synthetic PCRTags were divided by the wild-type ones in BY4741 and compared with the ratio of RROs between PCRTags in BY4741 and synXV_3.1.