Figure 1.
Chromatin modifications display lineage-specific associations with gene expression
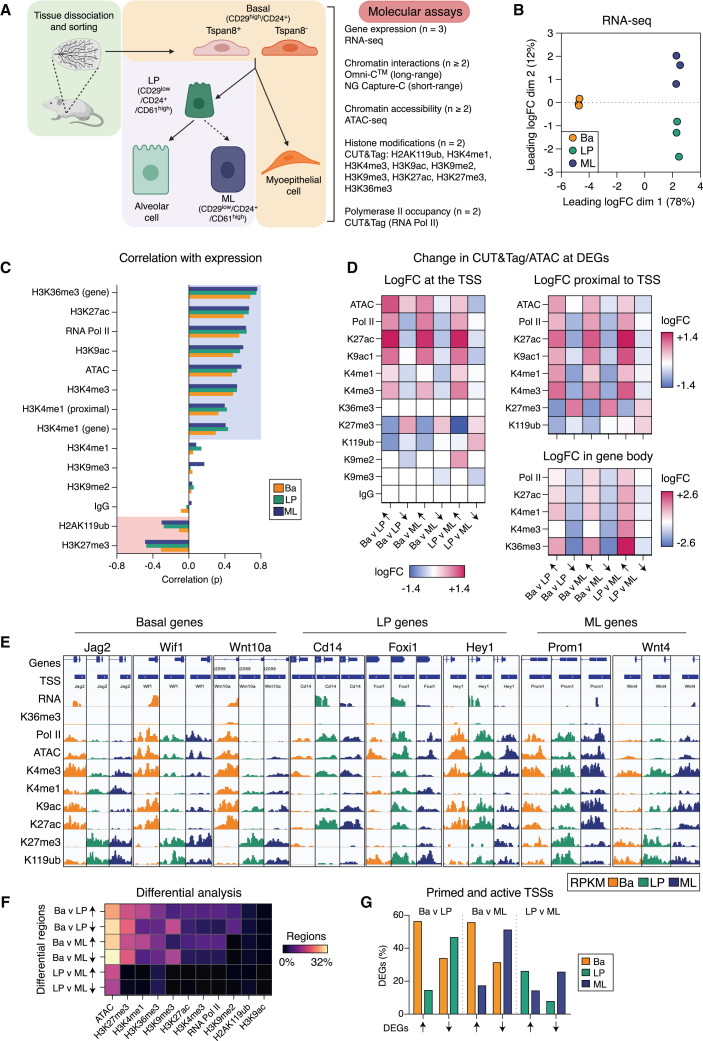
(A) Experimental design for the mammary epithelial epigenetic atlas encompassing basal, LP, and ML cells as well as Tspan8+ (quiescent) and Tspan8− (activated) basal cells.
(B) Multidimensional scaling (MDS) plot of RNA-seq data.
(C) Pearson correlation coefficients (p) for each epigenetic mark measured by CUT&Tag and ATAC-seq versus RNA expression.
(D) Heatmap of chromatin changes for DEGs between cell types for ATAC-seq and CUT&Tag (reads per kilobase per million [RPKM], log2) data.
(E) Coverage track plots for ATAC-seq and CUT&Tag at gene promoters.
(F) Heatmap showing the percentage of regions that display significant differential logFC (>0 up/<0 down) for ATAC-seq and CUT&Tag marks between the three cell types (FDR < 0.05).
(G) Bar plot showing TSSs considered to be primed or active across DEGs. See also Figures S1 and S2; Tables S1, S2, and S3.