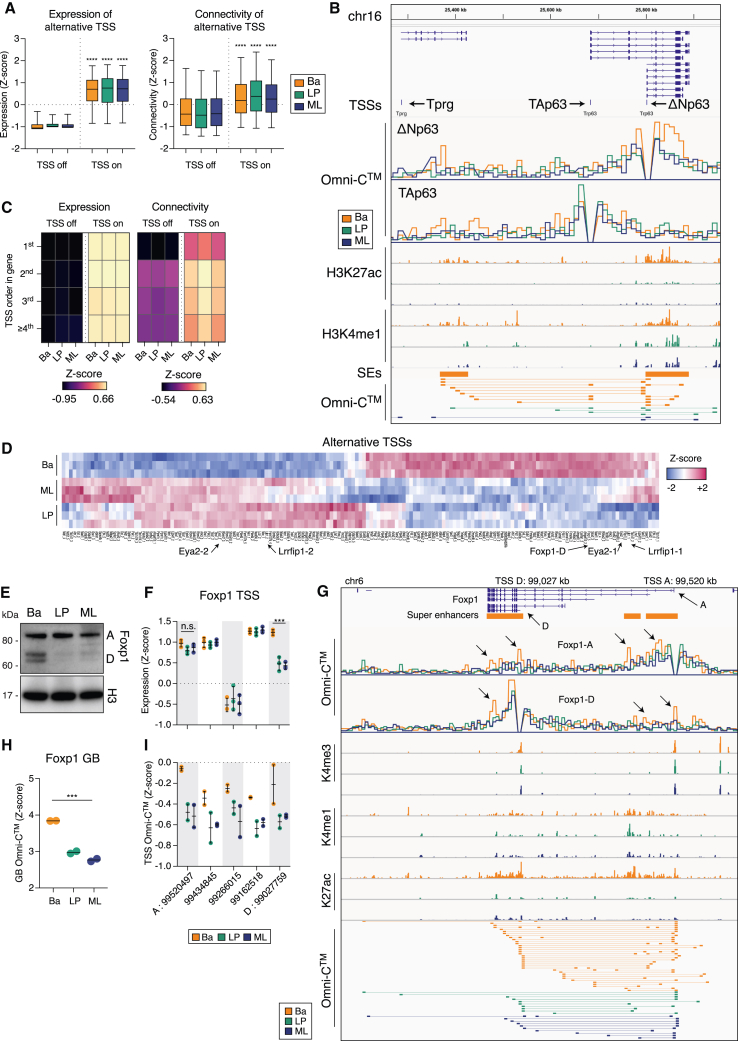
Figure 4.
Super-enhancer interactivity determines Trp63 and Foxp1 isoform expression
(A) Expression and connectivity of TSSs identified as “off” or “on.” t test with Welch’s correction for “off” versus “on” TSSs. ∗∗∗∗p < 0.0001. Boxes shows median, quartiles, and 5th and 95th percentiles.
(B) Coverage track plot of the Trp63 locus. Shown are the isoforms, alternative TSSs, in silico 4C for ΔNp63 and TAp63 TSSs (CCPM), CUT&Tag (RPKM), SEs, and interactions between the TSSs and the distal regions.
(C) Heatmaps of expression and chromatin connectivity for TSSs based on their position within the gene.
(D) Heatmap of DEATSS expression with examples indicated.
(E) Western blot for Foxp1 expression. Histone 3 (H3) was used as a loading control.
(F) Foxp1 TSS expression, annotated as genomic coordinates.
(G) Chromatin state of the Foxp1 locus. Shown are the isoforms, SEs, in silico 4C (CCPM) for Foxp1-A and Foxp1-D TSSs, coverage (RPKM) for CUT&Tag, and Omni-C interactions called by FitHiChIP.
(H) Foxp1 gene-body (GB) connectivity determined by in silico 5C analysis.
(I) Foxp1 chromatin interactivity between the alternative TSSs and the GB determined by in silico 4C analysis. Genome coordinates are from Mus musculus mm10 (GRCm38). The p values were determined using one-way ANOVA: ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S4; Table S7.