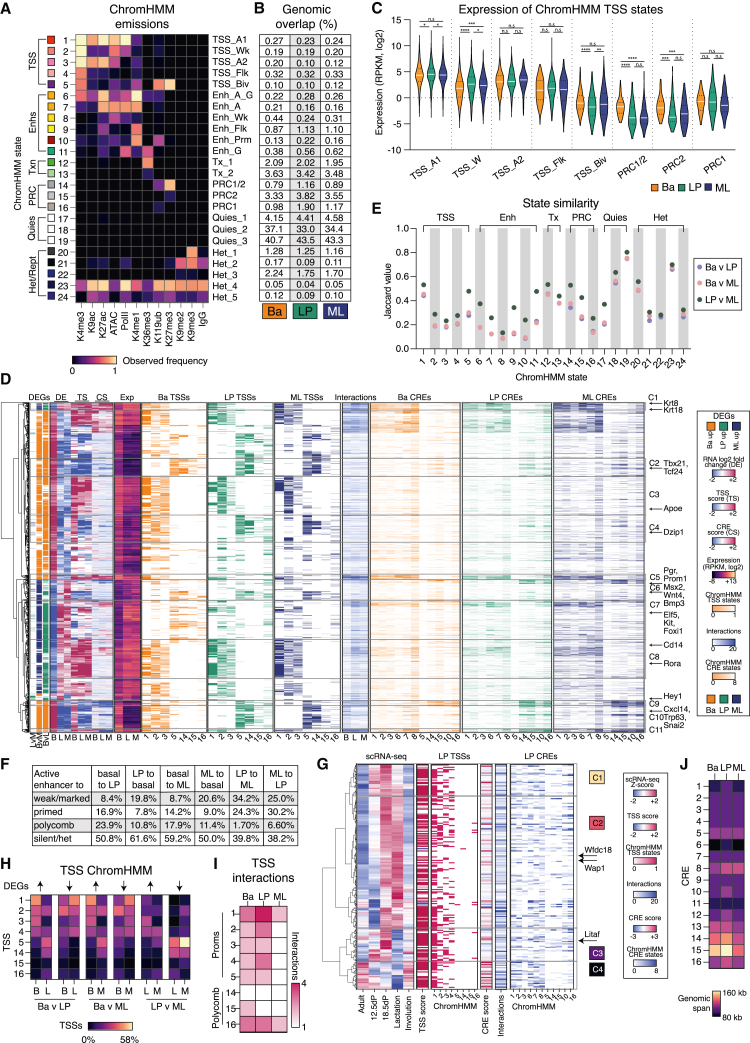
Figure 5.
Chromatin state modeling reveals differential usage of enhancers and polycomb regions in the different mammary epithelial lineages
(A) Epigenomes of epithelial subsets modeled as 24 chromatin states with ChromHMM using ATAC-seq and CUT&Tag data. Heatmap displaying frequencies of chromatin marks and IgG (control) across each state, with annotations listed on the left and terms on the right. A1/2, active; Biv, bivalent; Flk, flanking; G, genic; Prm, primed; Wk, weak.
(B) Genomic overlap of ChromHMM states across MECs.
(C) Expression of genes with a defined TSS, states 1–5, and polycomb states 14–16. The p values were determined using two-tailed t test with Welch’s correction: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(D) Hierarchical clustering of DEGs by expression, TSS, and enhancer score. Shown from left to right is the cluster dendrogram, DEGs for pairwise comparisons, differential expression log2 FC (DE), TSS score (TS), CRE score (CS), expression (RNA-seq, RPKM, log2), TSS ChromHMM states, number of significant interactions as determined by FitHiChIP, CRE ChromHMM states, cluster numbers, and highlighted genes.
(E) State similarity (Jaccard values) for pairwise comparisons of basal, LP, and ML cells.
(F) Active enhancer regions (states 6 and 7) displayed as altered state for each pairwise comparison. Shown is the percentage of active enhancer regions in one subset and the percentage of those same regions that are weak/marked (state 8) or primed (state 10), polycomb only (states 14, 15, and 16), or silent/heterochromatin (states 17 to 24) within the comparator subset.
(G) Hierarchical clustering of LP-alveolar genes, as identified in Pal et al.25 Shown from the left to right: cluster dendrogram, single-cell RNA expression (Z scores), TSS score, TSS ChromHMM states contributing to the TSS score, CRE score, number of significant interactions as called by FitHiChIP, CRE ChromHMM states contributing to the CRE score, subclusters, and highlighted genes.
(H) Chromatin state for TSS regions of DEGs.
(I) The number of interactions detected by FitHiChIP for genes categorized based on their TSS chromatin state.
(J) The median genomic distance between TSSs and CREs. See also Figures S5, S6, and S7; Table S8.