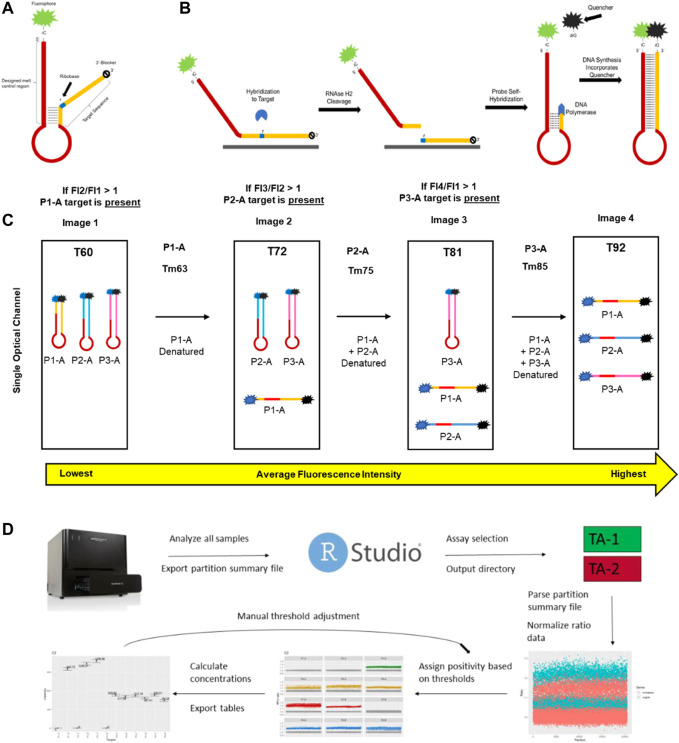
FIGURE 1.
Key aspects of mdPCR. (A) Melt-based probe design. (B) The probe binds to its amplicon target, causing the formation of an RNA:DNA duplex at the ribobase (r) present in the probe. RNAse H2 present in the reaction mixture cleaves the probe, allowing the 5′ portion of the cleaved probe to form a hairpin which is extended by DNA polymerase. During extension, the DNA polymerase directs a site-specific incorporation of a fluorescent quencher to form a temperature-reversible quenched hairpin. (C) Post-amplification, probes containing the same fluorophore can be discriminated based on the hybridization temperature of the final hairpin as determined by discrete melt analysis in which the fluorescence is measured by imaging at four temperatures and the ratios of each set of temperatures (T2/T1, T3/T2, and T4/T3) are used to determine presence/absence of each target. (D) Workflow for Erebos data analysis. Optimized positivity thresholds (ratios) for each probe (previously determined experimentally) used were: P1-A > 1.075, P1-B > 1.1, P2-A > 1.06, P2-B > 1.11, P3-A > 1.08, P3-B > 1.12, P4-A > 1.07, P4-B > 1.1, P5-A > 1.085, P5-B > 1.1, P6-A > 1.08 and P6-B 1.075. The copies per μL, total copies per assay input, allele frequency, and 95% Poisson confidence intervals were calculated for each target.