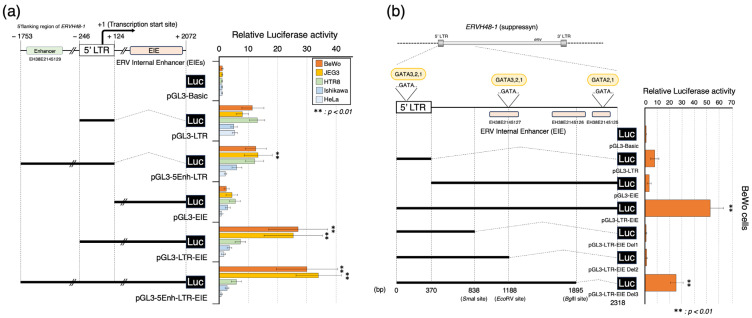
Figure 4.
Luciferase activities of the ERVH48-1 promoter and novel enhancer sequences (EIEs). (a) The schematic diagram on the left depicts the suppressyn 5′ LTR and its flanking sequences, including the 5′ flanking region of ERVH48-1 and putative enhancer sequences (ERV Internal Enhancers; EIEs). Below, the separate constructs cloned into the pGL3 vector and the regions they cover, including the 5Enh, LTR, and EIE sequences, are shown. On the right, relative luciferase activity was plotted for these constructs for each cell line used (BeWo, JEG3, HTR8, Ishikawa, and HeLa cells). (b) The left side of this diagram depicts a schematic of the ERVH48-1 5′ LTR and EIE sequences. Details of the whole and the restriction enzyme-truncated (SmaI, EcoRV, and BglII) pGL3 constructs are shown below this. Three enhancer regions in the EIEs were predicted via cCRE. Predicted binding sites for GATA transcription factors in this region are presented (…GATA…). Luciferase activity associated with the transfection of these constructs into BeWo cells is shown on the right. Luciferase activity associated with the presence of the pGL3-basic construct was used as a comparator for statistical analysis. Mann–Whitney U tests were performed, and statistical significance was set at ** p < 0.01. Experimental replicates were performed in duplicate, and the means ± SDs were calculated from three independent experiments.