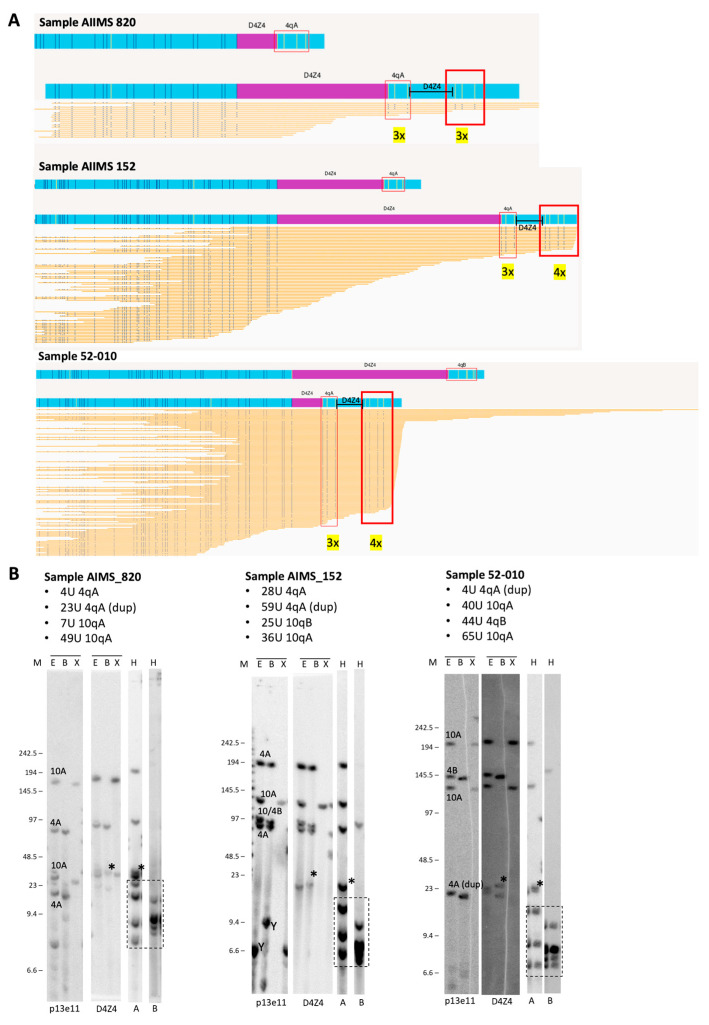
Figure 4.
(A) Duplicated 4qA signatures identified via visual inspection of OGM data using EnFocus FSHD analysis browser view (version 1.0; Bionano Access Software v1.7) for the analysis of probands AIIMS 820, AIIMS 152, and 52-010. The OGM output files show that all reads aligned to the D4Z4 alleles on chromosome 4. Nick sites are denoted by the red box, and the number of sites is given in yellow. (B) Comparison with SB analysis shows hybridisations with probes p13E-11 and D4Z4 on genomic DNA digested using the enzymes EcoRI/HindIII (E), EcoRI/BlnI (B), and XapI (X) and hybridisations with probes A and B on genomic DNA digested with HindIII (H). The alleles based on the different blots are indicated in the p13E-11 blot. The chromosome Y fragment is indicated, and the cross-hybridising fragments in the A/B hybridisations are marked with a dotted box. The size of the molecular weight marker is indicated on the left, and the asterisk (*) indicates the extra allele that appears upon D4Z4 and 4qA hybridisation and depicts the duplication allele. Original blot images can be found in Figure S1.