Abstract
An immunomagnetic separation (IMS) technique was developed to facilitate selective isolation of Mycobacterium paratuberculosis cells from milk. Rabbit polyclonal antibodies against radiation-killed intact M. paratuberculosis cells were produced and used to coat sheep anti-rabbit immunoglobulin G (IgG) type M-280 Dynabeads. The rabbit anti-M. paratuberculosis IgG-coated beads (IMB) reacted strongly with laboratory strains of M. paratuberculosis as determined by slide agglutination, and microscopic examination confirmed that M. paratuberculosis cells attached to the IMB. The IMB were found to have a maximum binding capacity of 104 to 105 CFU of M. paratuberculosis. Studies showed that IMS selectively recovered M. paratuberculosis from inoculated milk containing as few as 10 CFU of M. paratuberculosis per ml when 10 μl of IMB (ca. 106 beads) was added to 1 ml of milk and the preparation was incubated for 30 min at room temperature with gentle agitation. Larger volumes of milk (10 and 50 ml) were centrifuged and resuspended in 1 ml of phosphate-buffered saline–0.05% Tween 20 prior to IMS in order to increase the sensitivity of the method. Currently, primary isolation of M. paratuberculosis from a milk sample relies on chemical decontamination, followed by culturing on Herrold’s egg yolk medium, which must be incubated at 37°C for up to 18 weeks. The potential value of our IMS method is as an aid for rapid detection of M. paratuberculosis in milk when it is used in conjunction with end point detection methods, such as IS900 PCR or an enzyme-linked immunosorbent assay.
Mycobacterium paratuberculosis causes paratuberculosis, commonly known as Johne’s disease, in cattle, sheep, goats, and other ruminants (2). Although not currently classified as a zoonotic agent, M. paratuberculosis has been identified in intestinal biopsy tissues from some patients with Crohn’s disease (CD) (1). CD is a chronic, incurable, low-grade inflammation of the terminal ileum, one of two similar diseases of the human gastrointestinal tract known as inflammatory bowel disease. Whether the presence of M. paratuberculosis in biopsy material indicates that this organism has a causative role in CD or is simply a complicating infection is still the subject of much debate. However, if M. paratuberculosis has a causative role in CD, then milk may be a possible vehicle of transmission of the organism from cattle to humans (7, 21). Detectable quantities of M. paratuberculosis have previously been found in the milk of both clinically infected (20) and subclinically infected (18, 19) cattle with Johne’s disease. One theory put forward to explain the increasing incidence of CD in humans in certain parts of the world is that the human population may be repeatedly exposed to low levels of M. paratuberculosis in the milk supply (7). This explains the interest in determining whether M. paratuberculosis is present in the general supply of fluid milk, both raw and pasteurized. Only one such study has been published to date. Millar et al. (13) used IS900 PCR to detect M. paratuberculosis in retail pasteurized cow’s milk in England and Wales and reported that overall, 7% of 312 milk samples tested positive for the presence of M. paratuberculosis DNA over a 19-month period. At peak periods up to 25% of the milk samples were positive as determined by IS900 PCR. However, the presence of viable cells was never confirmed by decontamination and culturing of PCR-positive milk samples, so the theory of repeated exposure of humans to viable M. paratuberculosis in milk was not substantiated by the results of this milk survey.
Determination of the incidence of M. paratuberculosis in milk supplies is fraught with difficulties. First, M. paratuberculosis is an extremely slow-growing organism which can take up to 20 weeks for primary isolation, whereas most other microorganisms in milk exhibit growth within 24 to 48 h. As no selective medium for M. paratuberculosis is available, successful isolation of M. paratuberculosis currently relies on selective suppression of nonmycobacterial contaminants in samples by chemical decontamination. The recommended decontamination procedure for M. paratuberculosis is treatment with 0.75% (final concentration) hexadecylpyridinium chloride (HPC) for several hours (23). A balance must be struck between adequate time for decontamination and the possibility of undue damage to the M. paratuberculosis cells if the decontamination period is too long. Unless adequate decontamination is achieved, any surviving undesirable microorganisms quickly overgrow the M. paratuberculosis colonies, thwarting isolation efforts. All of the milk surveys carried out to date (13, 18–20) have relied on chemical decontamination in some shape or form prior to culturing of M. paratuberculosis from milk. Second, M. paratuberculosis is likely to be present in low numbers in naturally infected milk samples. A titer of just 2 to 8 CFU of M. paratuberculosis per 50 ml of milk has been reported for milk obtained aseptically from asymptomatic cattle with Johne’s disease (19). Consequently, the culture methods employed to isolate M. paratuberculosis must be extremely sensitive, or, alternatively, the sensitivity of the culture method employed must be improved by concentrating the M. paratuberculosis cells prior to culturing. In theory, immunomagnetic separation (IMS) could be used to resolve these difficulties.
IMS is a simple but powerful method for extracting a desired organism from heterogeneous bacterial suspensions, such as those that are encountered in food, clinical specimens, and feces (3, 15). It has previously been used successfully with several types of food samples, including milk (8, 14, 16, 17). IMS relies on the interaction between cell surface antigens and antibodies attached to paramagnetic beads. The desired cells are separated by placing a bead suspension in a strong magnetic field. The beads can be resuspended after IMS in a smaller volume of liquid, thereby concentrating the sample. If appropriate antibodies directed against surface antigens of M. paratuberculosis were obtained or produced, this organism could be selectively isolated from milk samples and concentrated by IMS, thereby improving the specificity and sensitivity of subsequent culture methods. During IMS the M. paratuberculosis cells would not be exposed to potentially damaging chemicals, as they are during traditional decontamination procedures, and, consequently, the physiological state of the cells would not be affected. Previous applications of IMS in mycobacteriology include detection of Mycobacterium avium in stool samples from AIDS patients (10) and detection of Mycobacterium tuberculosis in cerebrospinal fluid (11).
In this paper we describe the development, optimization, and evaluation of an IMS procedure to facilitate the isolation of M. paratuberculosis from milk samples.
MATERIALS AND METHODS
Production of rabbit anti-M. paratuberculosis antiserum.
Colonies of M. paratuberculosis B4 (a bovine field strain isolated in Northern Ireland) grown on Herrold’s egg yolk medium (HEYM) were suspended in phosphate-buffered saline (PBS), and the preparation was centrifuged, washed five times in PBS, and then irradiated (dose, 15 kGy) with a Gammabeam 650 instrument (Nordion International Inc., Kanata, Ontario, Canada) in order to kill the cells. A 0.5-ml portion of a dense suspension (concentration, approximately 108 CFU/ml) of irradiated cells was mixed with an equal volume of the adjuvant Quil A (125 μg per ml; Superfos, Vedbaek, Denmark) and used to inoculate a rabbit subcutaneously at multiple sites. This inoculation procedure was repeated after 5 weeks, followed (at 15- to 20-day intervals) by three intravenous inoculations of 0.5 ml. The rabbit was test bled 7 to 14 days after the second subcutaneous inoculation and after each intravenous inoculation. The antiserum obtained was tested by an enzyme-linked immunosorbent assay (ELISA) in which microtiter wells were coated with the cell suspension used to inoculate the rabbit; HEYM was used as a control. No ELISA reaction was obtained with the HEYM antigen. The rabbit was exsanguinated 14 days after a titer greater than 1:3,000 was recorded with the M. paratuberculosis antigen; the serum was separated and stored at −20°C.
Purification of the polyclonal rabbit anti-M. paratuberculosis serum.
The rabbit anti-M. paratuberculosis serum was purified by precipitating albumin and other non-immunoglobulin G (IgG) proteins with caprylic acid (25 μg per ml) after 2 volumes of 0.06 M acetate buffer (pH 4.3) was added by the method of McKinney and Parkinson (12). After centrifugation at 10,000 × g for 15 min, the precipitate was discarded, and the IgG fraction was dialyzed overnight at 4 to 10°C against 0.01 M PBS (pH 7.2). The purified polyclonal IgG was divided into 1-ml aliquots and stored at −20°C.
Specificity of the polyclonal rabbit anti-M. paratuberculosis IgG.
Slide agglutination was used to test for cross-reactions of the polyclonal IgG with other Mycobacterium spp. and bacterial isolates obtained from raw milk. Twenty microliters of undiluted polyclonal IgG was applied to a clean slide. A loopful of the test organism was mixed with the IgG, the slide was tilted several times, and agglutination was checked within 2 min. The slide agglutination test was repeated by using 20-μl portions of 1:10, 1:100, and 1:1,000 dilutions of the polyclonal IgG in PBS.
Coating of magnetic beads.
The polyclonal IgG was used to coat sheep anti-rabbit IgG type M-280 Dynabeads (catalog no. 11203; Dynal UK Ltd., Wirral, United Kingdom) for 24 h at 2 to 4°C as recommended by the manufacturer. In all of the IMS trials 10-μl aliquots (ca. 106 beads) of the rabbit anti-M. paratuberculosis IgG-coated beads (IMB) were employed.
Strains studied.
Three M. paratuberculosis strains were used in this study. These strains were type strain NCTC 8578 and field strains B2 and B4, which were previously isolated from cattle in Northern Ireland. The culture conditions used and the method used to prepare the inoculum have been described previously (6). In addition, Mycobacterium intracellulare NCTC 10425, Mycobacterium kansasii NCTC 10268, and a field strain of M. avium were used to test the specificity of the polyclonal IgG.
Milk samples.
Milk samples with low levels of background microflora (either raw cow’s milk aseptically obtained from a healthy Friesian cow as described previously [6] or pasteurized whole cow’s milk routinely sent to the Food Microbiology Unit for testing), were used in this study.
Confirmation of M. paratuberculosis attachment to IMB.
In order to visualize M. paratuberculosis cells bound to the IMB after IMS, 2 drops of a bead-cell suspension was transferred to a microscope slide, air dried, and then heat fixed. The smear was flooded with a 0.1% (wt/vol) phenolic auramine O solution for 15 min, decolorized with acid-alcohol for 2 min, counterstained with a 0.3% (wt/vol) methylene blue solution for 2 min, and then allowed to air dry prior to microscopic examination under blue light (9). Acid-fast cells fluoresced bright yellow against a dark background, whereas the IMB fluoresced only weakly.
Determination of the optimum immunocapture time and percentage of recovery.
Seven 1-ml aliquots of milk and three 1-ml aliquots of PBS were each inoculated with 106 CFU of M. paratuberculosis NCTC 8578. One of the inoculated milk samples was immediately serially diluted and cultured on HEYM slopes in order to determine the number of M. paratuberculosis cells added to the samples prior to IMS. Another three inoculated milk samples were centrifuged (2,500 × g for 15 min) and resuspended in 1 ml of PBS before IMS; these samples were designated milk/PBS samples. Ten microliters of coated Dynabeads was added to each inoculated sample (three milk samples, three milk/PBS samples, and three PBS samples), and the tubes were incubated on a Dynal sample mixer at room temperature (21°C) for 15, 30, or 60 min. After a 15-min immunocapture, one milk sample, one milk/PBS sample, and one PBS sample were removed and processed by using the remainder of the IMS procedure. Following IMS, the IMB were resuspended in 1 ml of PBS. After 30- and 60-min immunocapture times, three additional tubes were removed and processed in the same way. All samples were then diluted as necessary, and the number of M. paratuberculosis cells recovered by IMS from each of the suspending media after each of the immunocapture times was determined by culturing on HEYM slopes. The percentage of recovery of M. paratuberculosis cells from each milk or PBS sample was calculated on the basis of the number of cells which attached to the IMB. The same experiment was performed with M. paratuberculosis B2.
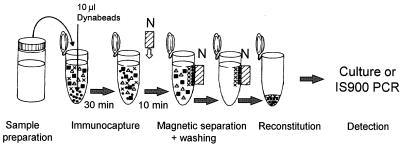
A schematic diagram of the optimized IMS procedure used for the remainder of this study is shown in Fig. 1. Briefly, 10 μl of IMB (106 IMB) was added to 1 ml of a test sample, and the preparation was incubated at room temperature with gentle agitation on a Dynal sample mixer for 30 min. After incubation the IMB were separated from the cell suspension with a magnetic particle concentrator (model MPC-M; Dynal) for 10 min. The residual liquid was removed by aspiration, and three washes in PBS containing 0.05% Tween 20 (PBS-T), with separation with the model MPC-M concentrator for 2 min between washes were performed. The IMB were resuspended in 1 ml of sterile water prior to culturing.
FIG. 1.
Schematic diagram of the optimized IMS procedure for detection of M. paratuberculosis in milk. N, magnet.
Later, the percentage of recovery by IMS was determined by counting the M. paratuberculosis cells lost with the milk after the magnetic separation step (i.e., the cells that were not able to bind to the IMB during the immunocapture step). A 10-fold dilution series of milk samples (1-ml aliquots) containing 106 to 102 CFU of M. paratuberculosis/ml was prepared. Each milk sample was subjected to IMS as described above. Following immunocapture and magnetic separation for 10 min, each milk sample was carefully aspirated into sterile plastic tubes, taking care not to dislodge any of the IMB. Each aspirate was diluted as necessary and cultured on HEYM slopes (100 μl per slope) to obtain a colony count for each milk sample. A 1-ml milk sample that was inoculated with 106 CFU of M. paratuberculosis but not subjected to IMS was simply diluted and inoculated onto HEYM slopes to confirm the number of M. paratuberculosis cells present in the most concentrated sample before IMS. Each successive 10-fold dilution of this milk sample was assumed to contain 10 times fewer M. paratuberculosis CFU than the previous dilution. This experiment was carried out on three separate occasions. The number of cells recovered by IMS was estimated by subtracting the number of M. paratuberculosis cells lost in the aspirate from the number of cells present in the milk sample prior to IMS. The percentage of recovery from each milk sample was calculated accordingly.
Use of centrifugation prior to IMS to concentrate M. paratuberculosis cells.
Different volumes of raw milk (5, 10, and 50 ml) were inoculated with a fixed number of M. paratuberculosis cells (approximately 106 CFU). Each milk sample was centrifuged at 2,500 × g for 15 min, and the pellet was resuspended in 1 ml of PBS-T prior to IMS. Following IMS, the IMB were resuspended in 1 ml of sterile water, and the number of M. paratuberculosis cells recovered from each milk sample volume was determined by serial dilution and inoculation of HEYM slopes (100 μl per slope). Colony counts were obtained after incubation of the slopes for up to 12 weeks at 37°C. This experiment was repeated twice.
In order to assess whether the majority of the M. paratuberculosis cells present in a milk sample were located in the pellet after centrifugation, two 10-ml samples of raw milk were inoculated with a fixed number of M. paratuberculosis cells (approximately 106 CFU) and centrifuged at 2,500 × g for 15 min in order to obtain the following three milk fractions: cream, whey, and pellet. The cream and pellet fractions were resuspended in 1 ml of PBS to facilitate enumeration of the M. paratuberculosis cells present. The whey fraction was tested directly. Following dilution as necessary, the number of M. paratuberculosis cells in each fraction was determined by inoculating HEYM slopes (100 μl per slope) and counting the colonies after incubation for up to 12 weeks at 37°C. The percentage of M. paratuberculosis cells in each fraction was calculated by taking into account the dilution of the cream and pellet layers and the volume of each milk fraction.
Determination of the minimum detection limit of the IMS method.
An inoculated milk sample containing 108 CFU of M. paratuberculosis/ml (the size of the inoculum was confirmed by dilution and plating on HEYM) was serially diluted in milk to obtain a set of eight milk samples containing between 101 and 108 CFU of M. paratuberculosis/ml. A 1-ml aliquot of each of the milk samples was then subjected to IMS and resuspended in 1 ml of PBS prior to inoculation onto HEYM slopes (100 μl per slope) without further dilution in order to determine the presence of any M. paratuberculosis cells recovered by IMS. This experiment was repeated four times.
Comparison of the recovery of a fixed number of M. paratuberculosis cells by culturing after centrifugation alone, centrifugation and IMS, and centrifugation and decontamination with 0.75% HPC.
In previous M. paratuberculosis milk surveys (18, 19) the workers centrifuged and decontaminated the milk samples with 0.75% HPC before preparations were cultured on HEYM slopes. A study was carried out to compare the numbers of M. paratuberculosis cells recovered by culturing after centrifugation and IMS and by culturing after centrifugation and HPC decontamination from different volumes of milk (1, 5, 10, and 50 ml) inoculated with approximately 106 CFU of M. paratuberculosis/ml. Culturing after centrifugation alone was included as a control. Control milk samples were centrifuged at 2,500 × g for 15 min, and each pellet was resuspended in 1 ml of sterile water. IMS milk samples were centrifuged and resuspended in 1 ml of PBS prior to IMS as described above. IMB were resuspended after IMS in 1 ml of sterile water. For decontamination, milk samples were centrifuged, resuspended in 5 ml of 0.75% HPC, and incubated at room temperature for 4 h with occasional shaking. HPC-treated samples were then centrifuged again, and each pellet was resuspended in 1 ml of sterile water. All of the samples were then diluted as necessary and inoculated onto HEYM slopes (100 μl per slope). The slopes were incubated for up to 12 weeks, and colony counts were obtained.
RESULTS
Confirmation of attachment of M. paratuberculosis to coated IMB.
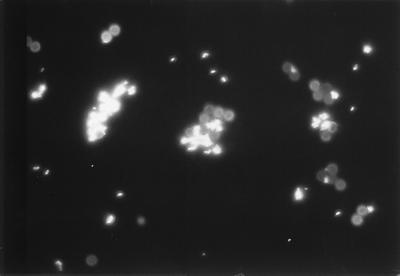
Fluorescent microscopy revealed that M. paratuberculosis cells attached to the IMB coated with purified rabbit anti-M. paratuberculosis IgG. It was evident that large clumps, not just single cells, were bound to some IMB, and often more than one cell or clump of cells were attached to the same IMB or group of beads (Fig. 2).
FIG. 2.
M. paratuberculosis cells attached to Dynabeads coated with polyclonal rabbit anti-M. paratuberculosis IgG and stained by the auramine O fluorescent acid-fast stain (bead diameter, 2.8 μm).
Specificity of purified rabbit anti-M. paratuberculosis polyclonal IgG.
The polyclonal IgG raised against M. paratuberculosis B4 produced strong agglutination reactions with all M. paratuberculosis strains available for testing in our laboratory (a total of 10 strains were tested, including 2 type strains and 8 field strains that originated from laboratories in the United States, Denmark, Northern Ireland, and Scotland). Moderate cross-reactions of the polyclonal IgG with three other Mycobacterium spp. and weak cross-reactions with bacterial isolates from raw milk were observed (Table 1). Table 1 also shows that a 1:1,000 dilution of the polyclonal IgG in PBS still reacted strongly with M. paratuberculosis NCTC 8578, whereas the levels of the cross-reactions with the other Mycobacterium spp. and the milk isolates diminished as the IgG was diluted.
TABLE 1.
Specificity of the rabbit anti-M. paratuberculosis polyclonal IgG as determined by slide agglutination
| Organism | Slide agglutination with the following dilutions of polyclonal IgG:
|
|||
|---|---|---|---|---|
| Undiluted | 1:10 | 1:100 | 1:1,000 | |
| M. paratuberculosis NCTC 8578 | ++a | ++ | ++ | ++ |
| M. avium (field isolate) | + | + | + | ± |
| M. intracellulare NCTC 10425 | ++ | + | + | ± |
| M. kansasii NCTC 10268 | ++ | ++ | ++ | + |
| Pseudomonas sp.b | + | ± | − | − |
| Staphylococcus sp.b | + | ± | − | − |
| Hafnia alveib | ± | − | − | − |
++, strongly positive; +, positive; ±, weakly positive; −, negative.
Isolates obtained from raw milk.
Determination of optimum immunocapture time.
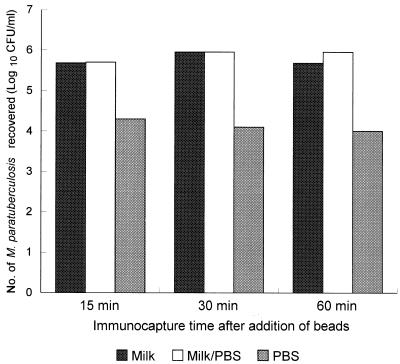
Immunocapture times of 15, 30, and 60 min (with gentle agitation) after addition of 10 μl of IMB were investigated. Similar trends were observed for the two M. paratuberculosis strains tested (NCTC 8578 and B2). Figure 3 shows how the number of M. paratuberculosis cells recovered by IMS was affected by immunocapture time and suspending medium. Overall, the numbers of M. paratuberculosis cells recovered by IMS from milk and milk/PBS samples were not significantly different, but the number of cells recovered from PBS alone was significantly less for both strains (P < 0.001). The percentage of recovery of M. paratuberculosis from each of the suspending media, which was calculated by determining the number of cells attached to the IMB, was surprisingly low. The highest percentage of recovery (37.1%) was obtained by incubating inoculated milk for 60 min with IMB, and the lowest percentage of recovery (≤0.2%) was obtained with inoculated PBS. The optimum immunocapture times for M. paratuberculosis cells varied with the suspending medium, as follows: whole milk, 30 to 60 min; milk-PBS, 30 min; and PBS, 15 min (Fig. 3). Subsequently, when the numbers of M. paratuberculosis cells which did not bind to the IMB were determined for a range of milk samples containing 102 to 106 CFU/ml, we found that the IMB had a maximum binding capacity of between 104 and 105 CFU (Fig. 4). Consequently, significant proportions of the inoculated M. paratuberculosis population were effectively lost when milk samples containing >104 CFU were subjected to IMS. This explains why just 37% of M. paratuberculosis cells were recovered by IMS from milk initially inoculated with 106 CFU/ml.
FIG. 3.
Optimum immunocapture times after addition of rabbit anti-M. paratuberculosis IgG-coated Dynabeads to inoculated milk, inoculated milk that was centrifuged and resuspended in PBS, and inoculated PBS.
FIG. 4.
Influence of the number of M. paratuberculosis cells present in milk on the percentage of recovery by IMS.
Effect of centrifugation prior to IMS.
M. paratuberculosis cells were present in all three milk fractions after centrifugation at 2,500 × g for 15 min. Overall, 13.0, 17.6, and 69.4% of the M. paratuberculosis cells present in a 10-ml milk sample segregated into the cream, whey, and pellet fractions, respectively, after centrifugation at 2,500 × g for 15 min. As the majority of the M. paratuberculosis cells were located in the pellet (P < 0.05), centrifugation was subsequently used to concentrate low numbers of M. paratuberculosis in larger volumes of milk prior to IMS. The numbers of M. paratuberculosis cells recovered by IMS after centrifugation of 5-, 10-, and 50-ml samples of milk inoculated with 106 CFU of M. paratuberculosis did not differ significantly (P > 0.05) (Fig. 5).
FIG. 5.
Comparison of the levels of recovery of M. paratuberculosis from inoculated milk by culturing on HEYM after centrifugation alone, after centrifugation followed by IMS, and after centrifugation followed by decontamination with 0.75% HPC.
Detection limit of the IMS method.
M. paratuberculosis was consistently isolated by the optimized IMS procedure from milk inoculated with 10 CFU/ml when milk samples inoculated with different concentrations of M. paratuberculosis (107 to 10 CFU/ml) were tested on four separate occasions. When centrifugation was used to concentrate the M. paratuberculosis cells in larger volumes of inoculated milk prior to IMS, the limit of detection by the culture method increased to 10 CFU per original volume (5, 10, or 50 ml) of milk tested.
Comparison of methods for the recovery of M. paratuberculosis from milk.
Similar numbers of M. paratuberculosis cells were recovered from milk samples by culturing after centrifugation alone and after centrifugation and HPC decontamination irrespective of the initial milk volume (Fig. 5). In contrast, significantly lower numbers of M. paratuberculosis cells were recovered by culturing after centrifugation and IMS (P < 0.001), although similar numbers of cells were recovered by this method from different volumes of milk. The latter finding is explained by the limited binding capacity of the IMB.
DISCUSSION
In this paper we describe successful development and optimization of a novel IMS method for isolation of M. paratuberculosis from milk. It was clear from previous IMS studies (3–5, 16, 17, 22) that IMS was not intended for quantification purposes since it was generally used with preenriched samples. Our novel IMS method for M. paratuberculosis isolation should aid in detection, not quantification, of this organism in milk when it is used in conjunction with a suitable end point detection method. The IMS method consistently detected M. paratuberculosis in milk samples inoculated with 10 CFU/ml when it was used in association with culturing on HEYM slopes. This level of detection was possible in the absence of selective preenrichment, which is commonly performed prior to IMS of other food pathogens (4, 5, 22), and therefore indicates that the method is sensitive. The optimum immunocapture time after addition of IMB varied depending on the nature of the medium in which the M. paratuberculosis cells were suspended. The longest immunocapture time (60 min) was that required for whole milk, and the shortest immunocapture time (15 min) was that required for PBS. Our experiments showed that when 10-ml volumes of inoculated milk were centrifuged, the majority (69.4%) of M. paratuberculosis cells segregated in the pellet. Therefore, in future studies involving this IMS method, we will centrifuge 50-ml volumes of milk, resuspend each pellet in 1 ml of PBS-T prior to IMS, and use an immunocapture time of 30 min in order to maximize the sensitivity of the method.
Research groups reporting new IMS methods generally assess the performance of an IMS method in terms of the percentage of recovery obtained with an inoculated population (5, 22). A similar approach was taken in this study, although we readily acknowledge that interpretation of CFU data for M. paratuberculosis is difficult because of the probability that CFU arise from both single cells and clumps of cells. Counts obtained after IMS are semiquantitative at best (3). Examination of bead-cell complexes after IMS by using fluorescent acid-fast staining clearly illustrated that several cells or clumps can be bound to each IMB or cluster of IMB (Fig. 2). Nevertheless, an indication of the capabilities of the new IMS method was obtained. Initially, the percentages of recovery by IMS from milk, milk/PBS, and PBS samples inoculated with 106 CFU of M. paratuberculosis/ml were calculated by counting the M. paratuberculosis cells attached to the IMB. The values obtained were disappointingly low compared with the values given in previously reported IMS studies performed with other food pathogens (5, 22). However, an alternative approach for determining the percentages of recovery of cells by IMS is to enumerate the cells which do not bind to the beads but are lost with the milk after the magnetic separation step. Milk samples inoculated with a range of M. paratuberculosis concentrations (106 to 102 CFU/ml) were examined by taking this alternative approach. The results indicated that 10 μl of IMB (approximately 106 beads) had a maximum binding capacity of 104 to 105 CFU. This meant that when 102 to 104 CFU of M. paratuberculosis was present in a 1-ml milk sample, the percentage of recovery was close to 100% (Fig. 4), whereas when >104 CFU was present in a 1-ml milk sample, a maximum of only 104 CFU could be recovered. Additional evidence that the maximum binding capacity is limited was provided by the finding that the concentrations of M. paratuberculosis recovered after centrifugation and IMS from various volumes of milk that were initially inoculated with 106 to 107 CFU of M. paratuberculosis were consistently around 104 CFU/ml, approximately 100 times lower than the concentrations obtained by culturing after centrifugation and HPC decontamination (Fig. 4). During this study, some batch variation in the binding capacity of the IMB was observed. For example, data in Fig. 3 indicate that the binding capacity approached 106 CFU, whereas data in Fig. 5, which were obtained with a different batch of Dynabeads, indicate that the maximum level of recovery was just over 104 CFU of M. paratuberculosis. The limited binding capacity of the coated IMB is of no real consequence since an accurate determination of the number of M. paratuberculosis cells present in a milk sample is not possible after IMS; in any case, high numbers of M. paratuberculosis would never be encountered in naturally infected milk. The novel IMS method for M. paratuberculosis used in conjunction with culturing gives an indication of the presence or absence of viable M. paratuberculosis cells.
We used polyclonal antibodies raised against radiation-killed M. paratuberculosis to coat magnetic beads in this study. Polyclonal antibodies are directed against a number of surface antigens rather than against a single surface antigen, which avoids the problem of the high level of specificity which can sometimes occur with monoclonal antibodies. It also increases the likelihood of isolating the desired organism. Unfortunately, since some of the surface antigens may not be unique to M. paratuberculosis, there is a possibility of nonspecific cross-reactions with the polyclonal IgG. Slide agglutination confirmed that there were some cross-reactions between the polyclonal IgG and other mycobacteria and milk bacteria (Table 1). We used undiluted polyclonal IgG to coat the sheep anti-rabbit IgG-coated Dynabeads and were generally able to isolate M. paratuberculosis from milk containing 10 CFU/ml initially, so even though the IMS method was not completely specific, it was very sensitive. We found that cross-reactions of the polyclonal IgG with the milk isolates, in particular, were eliminated by diluting the IgG (Table 1). Therefore, it may be possible to improve the specificity of the IMS method by coating magnetic beads with a dilution of the polyclonal IgG (1:100 or 1:1,000). However, using a lower concentration of polyclonal IgG to coat magnetic beads may reduce the sensitivity of the IMS method, and this possibility should be investigated. It may be possible to circumvent this shortcoming in terms of specificity by using IMS in conjunction with BACTEC radiometric medium, to which PANTA antibiotic supplement could be added to combat the growth of contaminants, rather than HEYM. Alternatively, instead of culturing to confirm the presence of M. paratuberculosis, IMS could be used in conjunction with IS900 PCR, which is specific for this organism.
IMS is a relatively simple procedure to perform, although it is a little laborious. However, there is a potential risk that IMB and, therefore, M. paratuberculosis cells may be lost during the washing steps if care is not taken when the supernatant is removed by aspiration between washes. Milk is notorious as a suspending medium which can create problems for IMS applications due to its high fat content (3). Particular care is needed when the supernatant is removed after the first 10 min of magnetic separation. At this stage the IMB tend to slip down the side of the tube as the supernatant is being removed by aspiration rather than being tightly captured on the wall of the tube. However, with each subsequent washing step as the IMB are cleaned and milk components trapped among the IMB are removed, the beads tend to become more tightly captured on the wall of the tube, and there is less likelihood that they will be accidentally discarded.
In summary, we developed and evaluated an IMS procedure for isolating M. paratuberculosis from milk samples. This IMS procedure takes less than 1 h, compared to at least 5 h for HPC decontamination prior to culturing, and was found to consistently result in isolation of M. paratuberculosis cells from milk containing 10 CFU/ml. Centrifugation of larger volumes of milk and resuspension in 1 ml of PBS-T prior to IMS considerably improve the sensitivity of the method. The potential value of this novel IMS method is not for quantification of M. paratuberculosis; rather, it can be used for rapid detection of this organism in milk when it is combined with end point detection methods, such as IS900 PCR or an ELISA (3). In future work we will concentrate on evaluating the use of this novel IMS method in conjunction with IS900 PCR as a rapid technique for screening milk and feces samples for the presence of M. paratuberculosis.
ACKNOWLEDGMENTS
This work was supported by funds from the Ministry of Agriculture, Fisheries and Food, United Kingdom.
We thank Bill Graham for assistance during irradiation of the M. paratuberculosis antigen and Colin Bell, who carried out the ELISA work.
REFERENCES
- 1.Chiodini R J. Crohn’s disease and the mycobacterioses: a review and comparison of two disease entities. Clin Microbiol Rev. 1989;2:90–117. doi: 10.1128/cmr.2.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocito C, Gilot P, Coene M, De Kesel M, Poupart P, Vannuffel P. Paratuberculosis. Clin Microbiol Rev. 1994;7:328–345. doi: 10.1128/cmr.7.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cudjoe K S, Patel P D, Olsen E, Skjerve E, Olsvik Ø. Immunomagnetic separation techniques for the detection of pathogenic bacteria in foods. In: Kroll R, Gilmour A, Sussman M, editors. New techniques in food and beverage microbiology. Oxford, United Kingdom: Blackwell Scientific Publications; 1993. pp. 17–29. [Google Scholar]
- 4.Cudjoe K S, Krona R, Olsen E. IMS: a new selective enrichment technique for detection of Salmonella in foods. Int J Food Microbiol. 1994;23:159–165. doi: 10.1016/0168-1605(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 5.Fratamico P M, Schultz F J, Buchanan R L. Rapid isolation of Escherichia coli O157:H7 from enrichment cultures of foods using an immunomagnetic separation method. Food Microbiol. 1992;9:105–113. [Google Scholar]
- 6.Grant I R, Ball H J, Neill S D, Rowe M T. Inactivation of Mycobacterium paratuberculosis in cow’s milk at pasteurization temperatures. Appl Environ Microbiol. 1996;62:631–636. doi: 10.1128/aem.62.2.631-636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermon-Taylor J. Causation of Crohn’s disease: the impact of clusters. Gastroenterology. 1993;104:643–646. doi: 10.1016/0016-5085(93)90438-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johne B, Jarp J, Haaheim L R. Staphylococcus aureus exopolysaccharide in vivo demonstrated by immunomagnetic separation and electron microscopy. J Clin Microbiol. 1989;27:1631–1635. doi: 10.1128/jcm.27.7.1631-1635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubica G P. Clinical microbiology. In: Kubica G P, Wayne L G, editors. The mycobacteria: a sourcebook, part A. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 133–175. [Google Scholar]
- 10.Li Z, Han Bai G, Von Reyn C F, Marino P, Brennan M J, Gine N, Morris S L. Rapid detection of Mycobacterium avium in stool samples from AIDS patients by immunomagnetic PCR. J Clin Microbiol. 1996;34:1903–1907. doi: 10.1128/jcm.34.8.1903-1907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazurek G H, Reddy V, Murphy D, Ansari T. Detection of Mycobacterium tuberculosis in cerebrospinal fluid following immunomagnetic enrichment. J Clin Microbiol. 1996;34:450–453. doi: 10.1128/jcm.34.2.450-453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney M M, Parkinson A. A simple, non-chromatographic procedure to purify immunoglobulins from serum and ascites fluid. J Immunol Methods. 1987;96:271–278. doi: 10.1016/0022-1759(87)90324-3. [DOI] [PubMed] [Google Scholar]
- 13.Millar D, Ford J, Sanderson J, Withey S, Tizard M, Doran T, Hermon-Taylor J. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows’ milk in England and Wales. Appl Environ Microbiol. 1996;62:3446–3452. doi: 10.1128/aem.62.9.3446-3452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramatsu Y, Maruyama M, Yanase T, Ueno H, Morita C. Improved method for preparation of samples for polymerase chain reaction for detection of Coxiella burnetii in milk using immunomagnetic separation. Vet Microbiol. 1996;51:179–185. doi: 10.1016/0378-1135(95)00203-0. [DOI] [PubMed] [Google Scholar]
- 15.Olsvik Ø, Popovic T, Skjerve E, Cudjoe K S, Hornes E, Ugelstad J, Uhlén M. Magnetic separation techniques in diagnostic microbiology. Clin Microbiol Rev. 1994;7:43–54. doi: 10.1128/cmr.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skjerve E, Olsvik Ø. Immunomagnetic separation of Salmonella from foods. Int J Food Microbiol. 1991;14:11–18. doi: 10.1016/0168-1605(91)90032-k. [DOI] [PubMed] [Google Scholar]
- 17.Skjerve E, Rorvik L M, Olsvik Ø. Detection of Listeria monocytogenes in foods by immunomagnetic separation. Appl Environ Microbiol. 1990;56:3478–3481. doi: 10.1128/aem.56.11.3478-3481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Streeter R N, Hoffsis G F, Bech-Nielsen S, Shulaw W P, Rings M. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am J Vet Res. 1995;56:1322–1324. [PubMed] [Google Scholar]
- 19.Sweeney R W, Whitlock R H, Rosenberger A E. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J Clin Microbiol. 1992;30:166–171. doi: 10.1128/jcm.30.1.166-171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor T K, Wilks C R, McQueen D S. Isolation of Mycobacterium paratuberculosis from the milk of a cow with Johne’s disease. Vet Rec. 1981;109:532–533. [PubMed] [Google Scholar]
- 21.Thompson D E. The role of mycobacteria in Crohn’s disease. J Med Microbiol. 1994;41:74–94. doi: 10.1099/00222615-41-2-74. [DOI] [PubMed] [Google Scholar]
- 22.Vermunt A E M, Franken A A J M, Beumer R R. Isolation of salmonellas by immunomagnetic separation. J Appl Bacteriol. 1992;72:112–118. doi: 10.1111/j.1365-2672.1992.tb01811.x. [DOI] [PubMed] [Google Scholar]
- 23.Whitlock R H, Rosenberger A E, Sweeney R W, Hutchinson L J. Culture techniques and media constituents for the isolation of Mycobacterium paratuberculosis from bovine fecal samples. In: Chiodini R J, Kreeger J M, editors. Proceedings of the Third International Colloquium on Paratuberculosis. International Association for Paratuberculosis; 1992. pp. 94–111. , Providence, R. I. [Google Scholar]
- 24.Widjojoatmodjo M N, Fluit A C, Torensma R, Keller B H I, Verhoef J. Evaluation of the magnetic immuno PCR assay for rapid detection of Salmonella. Eur J Clin Microbiol Infect Dis. 1991;10:935–938. doi: 10.1007/BF02005447. [DOI] [PubMed] [Google Scholar]
- 25.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]