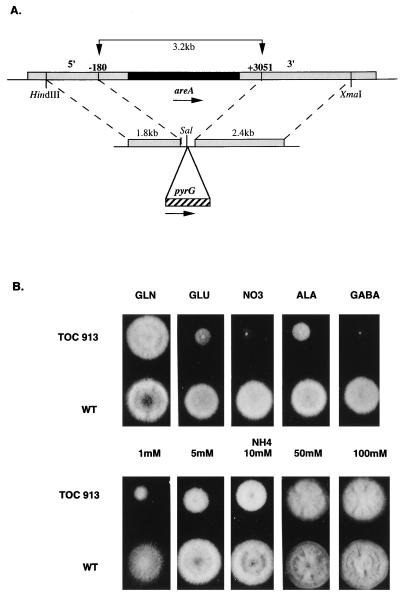
FIG. 3.
Inactivation of the A. oryzae areA gene. (A) Diagram of the construct used to inactivate the genomic A. oryzae areA gene. The shaded regions represent the 5′ and 3′ regions of the areA gene, and the solid region represents the areA coding sequences. In the inactivation construct the selectable marker pyrG (cross-hatched region) was flanked by sequences derived from the 5′ and 3′ ends of the areA gene (see text). This construct was transformed into a ΔpyrG recipient, and PyrG+ transformants were selected. The transformants were screened for the ability to use a variety of nitrogen sources, putative areA inactivation mutants were isolated, and inactivation of the areA gene was confirmed by Southern blot analysis. (B) Growth of the wild-type strain (WT) and ΔareA::pyrG strain TOC913 when glutamine (GLN), sodium glutamate (GLU), sodium nitrate (NO3), alanine (ALA), and GABA (final concentration of each compound, 10 mM) were used as sole nitrogen sources in glucose minimal media. Growth on ammonium tartrate (NH4) was also tested by using a range of concentrations, as indicated. Growth was scored after 2 days of incubation at 37°C.