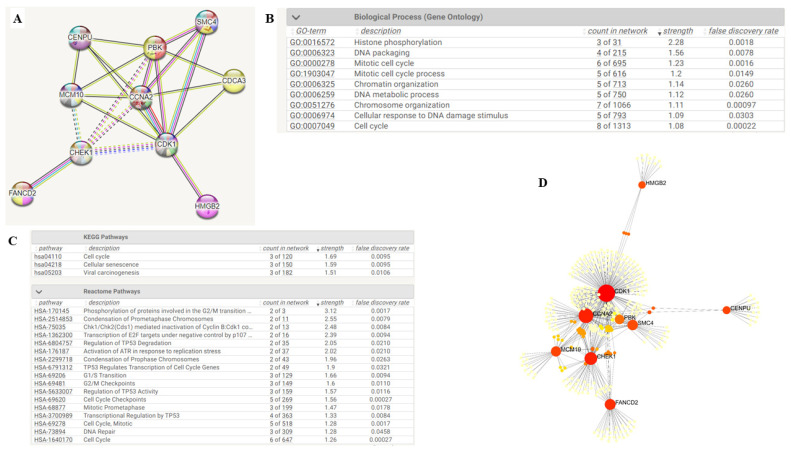
Figure 2.
Clustered PPI networks comprising CDK1/PBK/CHEK1 oncogenes in GBM and their functional enrichments. (A) A PPI cluster with a minimum interaction score significance > 0.700, 10 nodes, and 22 edges with an average local clustering coefficient of 0.82 and a PPI enrichment p-value of 2.11 × 10−15. The PPI network is from co-expression, text mining, databases, experiments, co-occurrence, neighborhood, and gene fusion. (B,C) show functional enrichments under the Gene Ontology (GO) biological processes, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome pathways that were available upon an analysis of the PPI network in the STRING database. (D) A signaling network cluster of KEGG enrichment analysis revealing CDK1/PBK/CHEK1 co-expression in the cell cycle analyzed in Network Analyst.