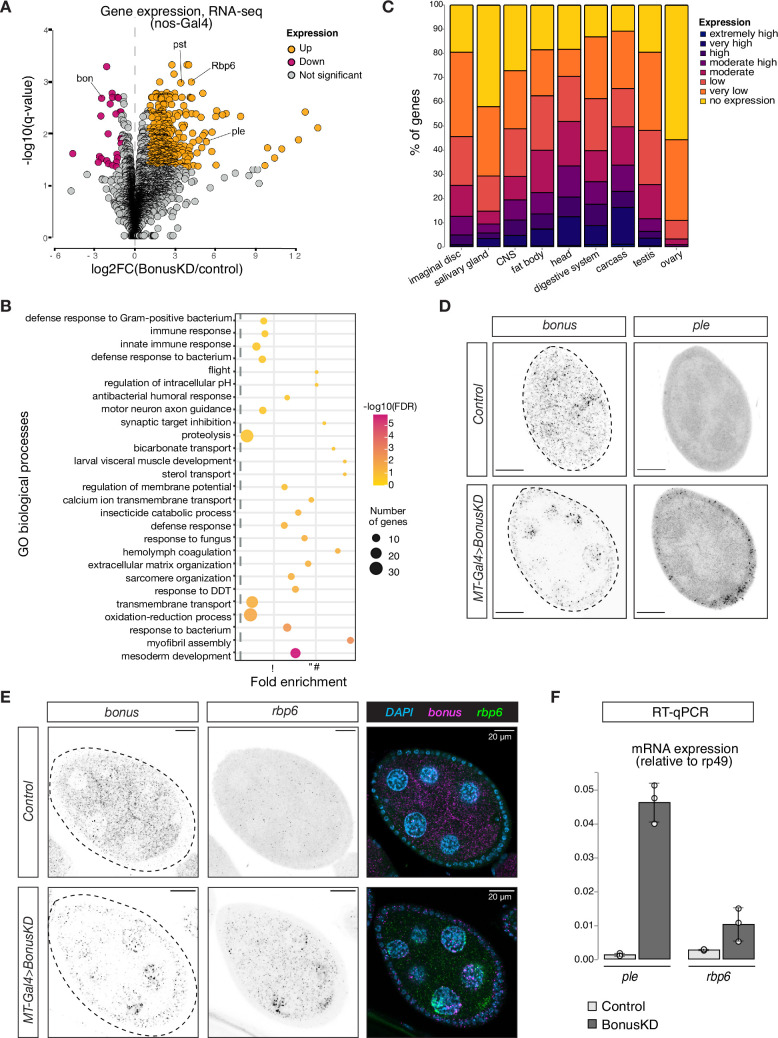
Figure 2. Bonus functions as a repressor of tissue-specific genes in ovary.
(A) Bon GLKD leads to misexpression of tissue-specific genes in the ovary. Volcano plot shows fold changes in genes expression upon Bon GLKD driven by nos-Gal4 in the ovary as determined by RNA-seq (n = 3). Siblings that lack shRNA against Bon produced in the same cross were used as a control. Genes that change significantly (log2FC >1, qval <0.05, LRT test, sleuth; Pimentel et al., 2017) are highlighted. Genes bon, pst, Rbp6, and ple are labeled. Genes with infinite fold change values (zero counts in control ovaries) are not shown. (B) Bon represses genes with diverse functions. Bubble plot shows the analysis of gene ontology (GO) enrichment at the level of biological processes (BP) for genes that are derepressed upon Bon GLKD driven by nos-Gal4 (log2FC >1, qval <0.05, LRT test, sleuth; Pimentel et al., 2017). Only GO terms above the established cut-off criteria (p-value <0.01 and >3 genes per group) are shown. BP are ranked by fold enrichment values. The most significant processes are highlighted in purple, and the less significant in yellow according to log10(FDR) values. The bubbles size reflects the number of genes, assigned to the GO BP terms. (C) Normal expression level of deregulated genes upon Bon GLKD in the tissues where they are normally expressed indicates Bon-mediated silencing of genes normally expressed in the head and digestive system. The graph shows the percentage of derepressed genes upon Bon GLKD driven by nos-Gal4 (log2FC >1, qval <0.05, LRT test, sleuth; Pimentel et al., 2017) with given expression level in the indicated enriched tissues. Expression levels according RPKM values from modENCODE anatomy RNA-seq dataset are no expression (0–0), very low (1–3), low (4–10), moderate (11–25), moderate high (26–50), high (51–100), very high (101–1000), and extremely high (>1000). (D) GLKD of Bon leads to ple expression in follicular cells. Confocal images of egg chambers show RNA in situ hybridization chain reaction (HCR) detecting ple and bonus mRNAs in flies with MT-Gal4>Bon GLKD and control siblings from the same cross that lack Bon shRNA (scale bar: 20 μm). (E) Bon represses rbp6 in the germline. Confocal images of egg chambers show RNA in situ HCR detecting rbp6 and bonus mRNAs in flies with MT-Gal4>Bon GLKD and control siblings from the same cross that lack Bon shRNA (scale bar: 20 μm). (F) Bar graph shows the relative expression of ple and rbp6 (normalized to rp49 level) in control and Bon-depleted ovaries (RT-qPCR, dots correspond to three independent biological replicates (n=3); error bars indicate st. dev.).