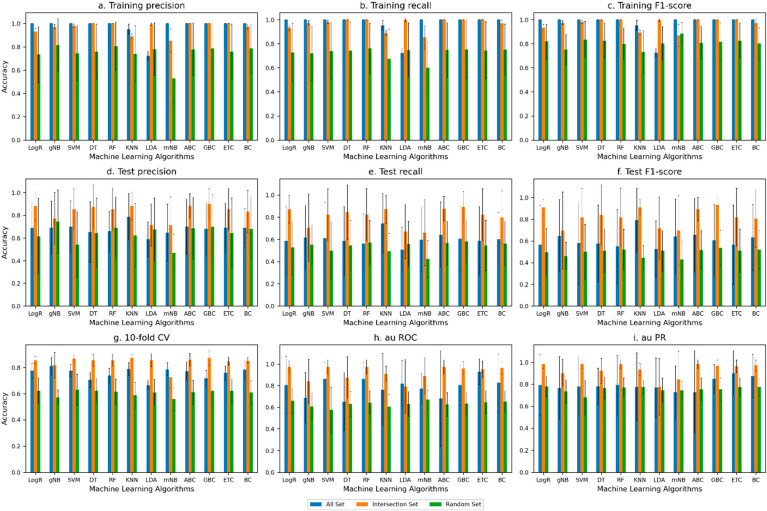
Figure 2.
Assessment of the performance of the machine learning algorithms in classifying multipartite and unipartite genomes based on gene-level analysis under 6-fold cross-validation setting; here, to begin with, all genes in the multipartite and unipartite genomes were considered. The performance metrics used were (a) training precision, (b) training recall, (c) training F1 score, (d) test precision, (e) test recall, (f) test F1 score, (g) 10f CV (ten-fold cross-validation), (h) AU ROC (area under ROC curve), and (i) AU PR (area under precision–recall curve). ‘All Set’ denotes all genes for training (as in the cross-validation partitioning), ‘Intersection Set’ refers to set of genes that consistently ranked high across all 6 rounds of cross-validation, and ‘Random Set’ refers to randomly sampled genes.