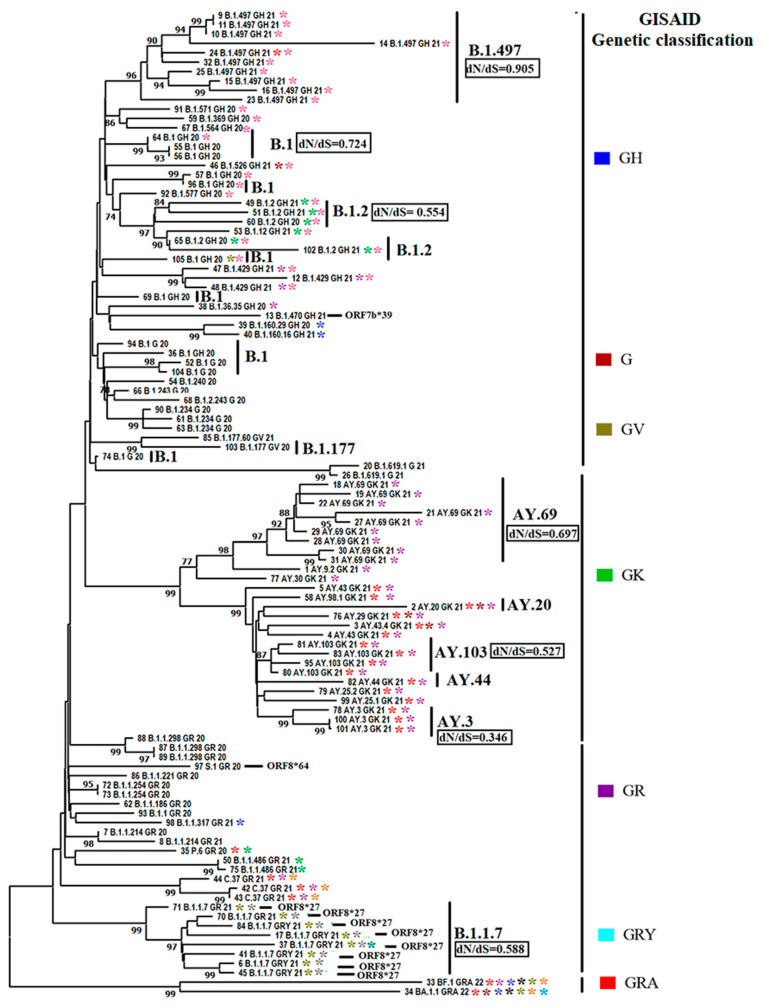
Figure 4.
Phylogenomics of SARS-CoV-2 lineages affecting cats. A phylogenetic analysis was conducted using 105 full-length viral sequences retrieved from the GISAID database (specific details about the sequences can be found in Supplementary File S1). The analysis was performed using the maximum likelihood method and the general reversible model. In the tree, labels B.1.497, B.1, B.1.2, B.1.177, AY.69, AY.20, AY.103, AY.44, AY.3, and B.1.1.7 identify clusters associated with the most prevalent SARS-CoV-2 pangolin lineages related to cat infections. The right pangolin lineages are grouped in the context of GISAID genetic group classification. The dN/dS ratios are indicated for the most prevalent cat lineages. Calculations were conducted using the FEL algorithm. Asterisks of different colors reflect specific codon sites at different genes evolving under positive selection in SARS-CoV-2 lineages affecting cats. Information about the codon sites associated with specific asterisk colors is presented in Figure 5 Black bars label lineages carrying premature stop codons at gen ORF8 codon positions 27 and 64.