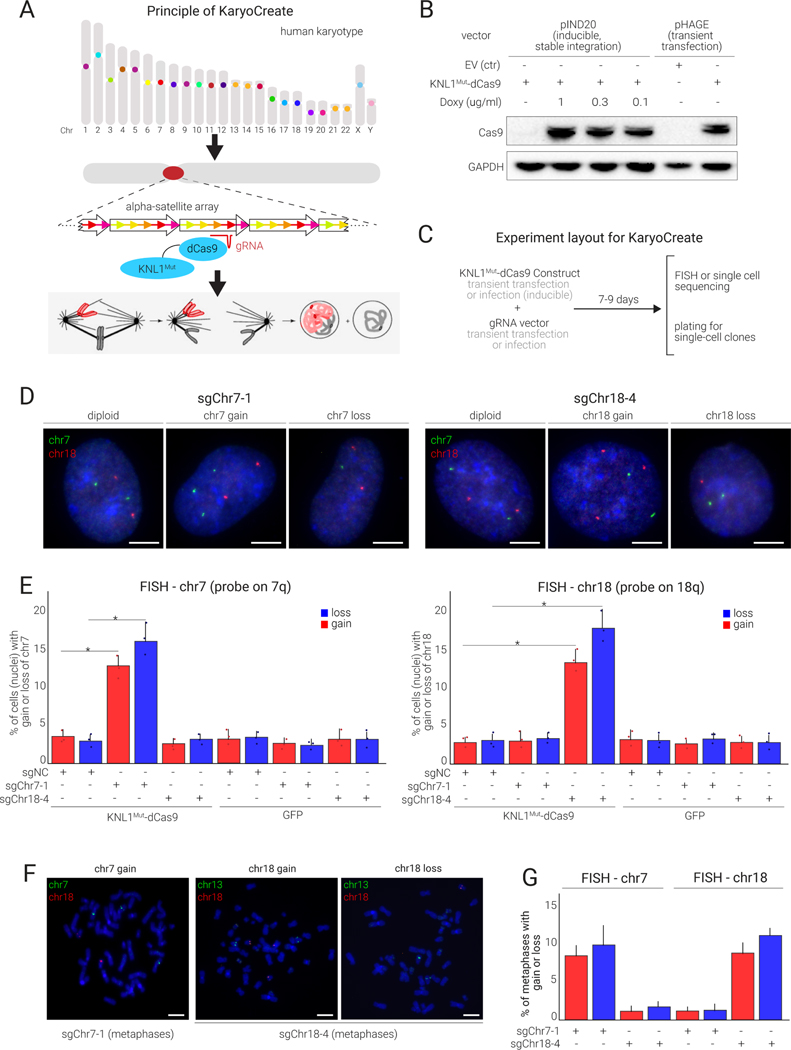
Figure 3.
KNL1Mut-dCas9 is recruited to human centromeres and allows induction of chromosome-specific gains and losses
(A) KaryoCreate conceptualization: Chromosome specificity of human α-satellite centromeric sequences makes it possible to induce missegregation of a specific chromosome while leaving the others unaffected.
(B) Western blot showing the expression of KaryoCreate constructs in hCECs, either through transient transfection with a constitutive promoter (pHAGE-CMV) or through infection with a doxycycline (Doxy)-inducible promoter (pIND20).
(C) KaryoCreate experimental plan with transient KNL1Mut-dCas9 expression and (transient or constitutive) sgRNA expression; cells are harvested after 7–9 days for validation by FISH and can then be plated to create single-cell clones.
(D) Representative FISH images using probes specific for chr7 or chr18 on hCECs showing gains and losses after KaryoCreate with the indicated sgRNAs.
(E) Quantification of the experiment shown in (D) for chr7 (left) or chr18 (right); see also Table S2 for automated image quantification. Mean and S.D. from triplicates.
(F) Representative metaphase spreads from hCECs treated as in (D) and analyzed by FISH using probes specific for chr7 and chr18 as indicated.
(G) Quantification of FISH signals from (F) (mean and S.D. from triplicates).