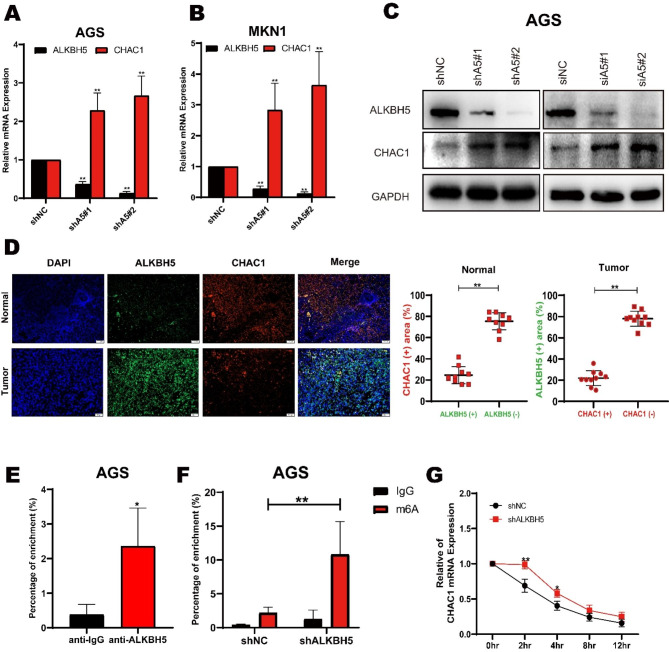
Fig. 5.
ALKBH5 regulates the expression of CHAC1 in GC. (A) Expression of two ALKBH5 knockdown sequences in AGS cells versus control for CHAC1 by RT-qPCR. (B) Expression of two ALKBH5 knockdown sequences in MKN1 cells was detected by RT-qPCR with CHAC1 expression in the control group. (C) Western blot detection of overexpression (left), sh knockdown (middle), and si knockdown (right) of CHAC1 and GCOM1 in AGS cells. (D) Correlation between ALKBH5 and CHAC1 protein expression in GC specimens., representative IF images of n = 10 GC specimens. Scale bar, Normal: 100 μm; Tumor: 50 μm. Fluoroscopic areas were calculated by Imaging J in 5 randomly selected microscopic fields for each cancer and paracancer specimen. t test calculates the percentage of ALKBH5 positive areas compared to ALKBH5 negative areas in CHAC1 positive areas; the percentage of CHAC1 positive areas compared to CHAC1 negative areas in ALKBH5 positive areas. Lines show the mean and standard deviation. (E) RIP (RNA immunoprecipitation) analysis of ALKBH5 enrichment of CHAC1 mRNA in AGS cells. (F) MeRIP-qPCR analysis of CHAC1 mRNA in AGS cells with or without ALKBH5 knockdown at m6A levels. (G) qPCR of CHAC1 mRNA stability in AGS cells with or without ALKBH5 knockdown. Identical amounts of RNA from cells treated with 2 μg/ml actinomycin D for 0 to 12 h were collected and measured by qPCR