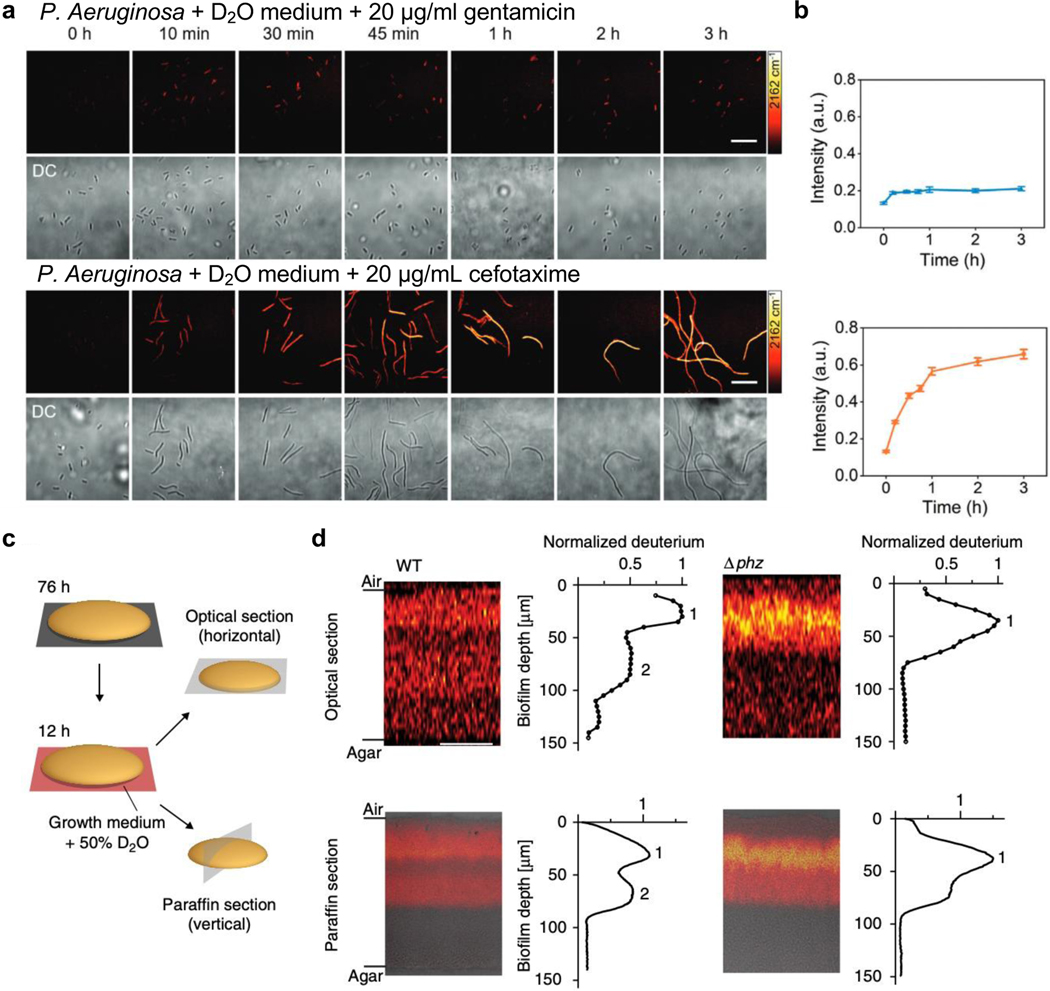
Figure 4. Assaying microbial metabolism with the biorthogonal SRS platform.
(a) Time-lapse SRS (targeting the C-D vibration, 2162 cm−1) and the corresponding transmission imaging of P. aeruginosa after culturing in D2O-containing medium with the addition of 20 μg mL−1 gentamicin (top) or cefotaxime (bottom). Scale bars: 20 μm. Adapted in part with permission from ref89, Copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) Corresponding average C–D intensity plot (designating labeled biomass) over time for P. aeruginosa after culturing in D2O-containing medium with gentamicin (top blue) or cefotaxime (bottom orange) treatment. Number of cells N ≥ 10 per group. Error bars: SEM. Adapted in part with permission from ref89, Copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) Schematic of two methods to visualize metabolic activity in colony biofilms. Optical sectioning uses the inherent 3D sectioning capability of SRS to acquire images in z-direction in 5 μm steps without sample preparation procedure. The paraffin section requires paraffin embedding and sectioning to provide 10-μm-thin slices for imaging. Adapted in part with permission from ref90, Copyright 2019 Nature publishing group. (d) SRS images and the corresponding plots of deuterium signals per biofilm depth for both optical sections and paraffin sections of colony P. aeruginosa biofilms after a 12-h incubation in D2O-containing medium. Data plots show the mean deuterium signal per biofilm depth. One replicate each of WT and Δphz is shown and is representative of at least five biological replicates. Scale bars: 50 μm. Adapted in part with permission from ref90, Copyright 2019 Nature publishing group.