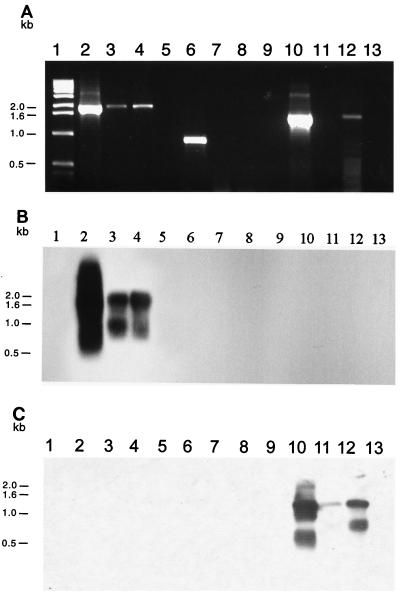
FIG. 5.
(A) Amplification of indigenous plasmid replication sequences from DNA extracted from marine sediment microbial communities: ethidium bromide-stained 0.8% agarose gel electrophoresis of PCR-amplified DNA obtained with either repSD41-specific primers (lanes 2 to 5), repSD164-specific primers (lanes 6 to 9), or repSD172-specific primers (lanes 10 to 13). Lane 1, 1-kb DNA size standard; lane 2, positive control (cesium chloride-purified plasmid pTM41 DNA); lane 3, total community DNA extracted from 1 g of sediment collected in November 1995; lane 4, total community DNA extracted from 1 g of sediment collected in June 1997; lane 5, negative control (sterilized sediment with no template DNA added); lane 6, positive control (cesium chloride-purified plasmid pTM164 DNA); lane 7, total community DNA extracted from 1 g of sediment collected in August 1995; lane 8, total community DNA extracted from 1 g of sediment collected in November 1995; lane 9, negative control (sterilized sediment with no template DNA added); lane 10, positive control (cesium chloride-purified plasmid pTM172 DNA); lane 11, total community DNA extracted from 1 g of sediment collected in November 1995; lane 12, total community DNA extracted from 1 g of sediment collected in June 1997; lane 13, negative control (sterilized sediment with no template DNA added). (B) Corresponding Southern blot analysis performed with the 1,302-bp repSD41 probe. (C) Corresponding Southern blot analysis performed with the 875-bp repSD172 probe. The band showing homology to the repSD172 probe below the predominant PCR product after 36 to 48 h of exposure to X-ray film represents smaller amplified replication products likely due to differential annealing of the PCR primers to the template DNA.