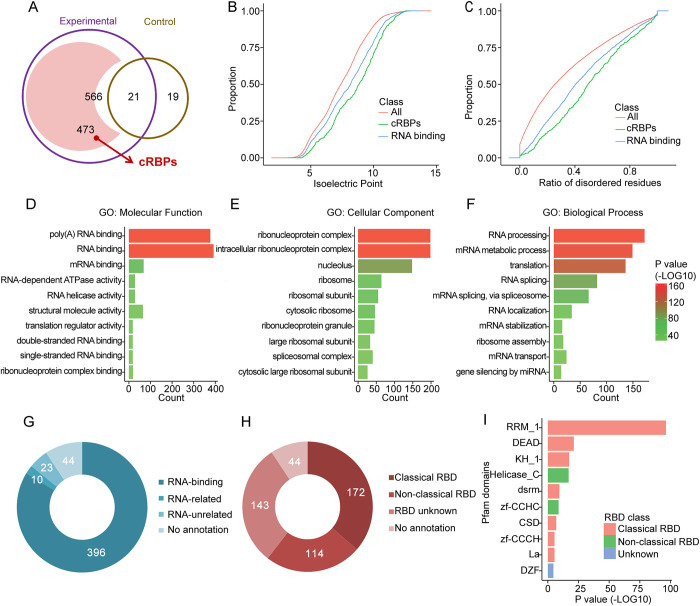
Fig. 2.
Characteristics of mRBPome-captured proteins in undifferentiated spermatogonia. (A) Venn diagram of oligo(dT)-captured proteins identified by mass spectrometry analysis in the UV-crosslinked (experimental) versus non-UV-crosslinked (control) samples (587 total candidates were obtained from three independent experiments, data are presented in Fig. 1E). After removing the 21 proteins that were also detected in control, 473 captured RBPs (cRBPs) were present in at least two biological replicates. (B,C) Distribution of isoelectric point and disordered residues among total proteins (magenta), undifferentiated spermatogonial mRBPome-captured proteins (blue) and known RBPs (green). (D-F) Significantly enriched Gene Ontology (GO) terms for mRBPome-captured proteins based on molecular processes (D), cellular components (E) and biological processes (F). (G,H) Classification of mRBPome-captured proteins according to the GO annotations of RNA-associated functions (G) and predefined domains (H). (I) Top ten enriched Pfam domains among mRBPome-captured proteins.