Figure 1.
Stag1 is required for naive pluripotency in mouse ESCs
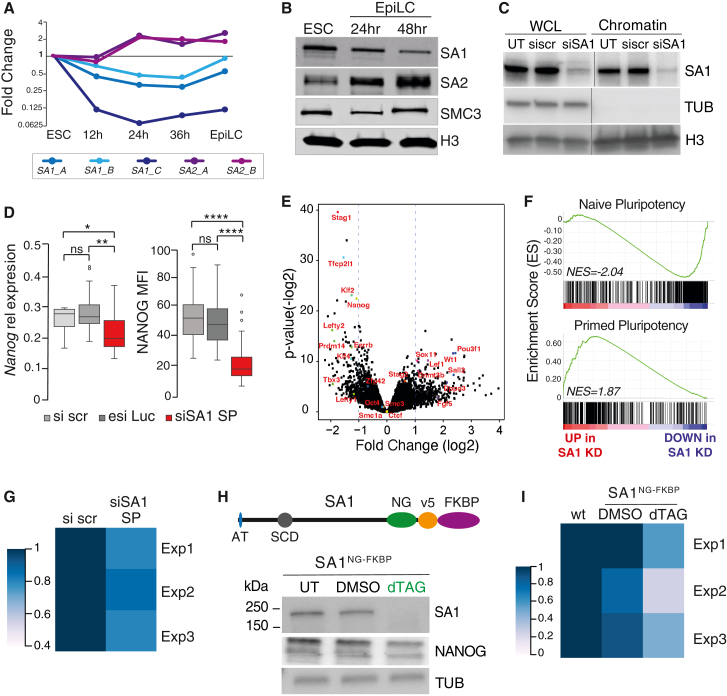
(A) Log2 fold change of Stag1 (SA1) and Stag (SA2) gene expression assessed by qRT-PCR during in vitro mESC differentiation toward EpiLCs. Multiple primer pairs were used for Stag1 (blue) and Stag2 (purple) mRNA (see box). Data are derived from two independent replicates.
(B) Whole-cell protein extracts (WCL) from naive mESCs and EpiLCs were analyzed by western blot (WB) for levels of SA1, SA2, and SMC3. H3 serves as a loading control.
(C) WB analysis of SA1 levels in WCL and chromatin fractions upon treatment with scrambled control siRNAs (si scr) or SmartPool SA1 siRNAs (siSA1) for 24 h in naive mESC cells. Tubulin (TUB) and H3 serve as fractionation and loading controls.
(D) Left: relative expression of Nanog mRNA by qRT-PCR in naive mESCs upon treatment with si scr, esiLuciferase control, or siSA1. Data are from eight independent replicates. Right, mean fluorescence intensity (MFI) of NANOG protein assessed by immunofluorescence (IF) in naive mESCs treated with same siRNAs as before. Cells were counterstained with DAPI. Data are n > 100 cells/condition across three independent replicates. Whiskers and boxes indicate all and 50% of values, respectively. The central line represents the median. Asterisks indicate a statistically significant difference as assessed using two-tailed t test. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001; ns, not significant.
(E) Volcano plot displaying the statistical significance (–log2 p value) versus magnitude of change (log2 fold change) from RNA-seq data produced in mESCs treated with si scr or siSA1 for 24 h. Data are from three independent replicates. Vertical blue dashed lines represent changes of 2-fold. Selected genes associated with cohesin, pluripotency, and differentiation have been highlighted in red.
(F) Enrichment score (ES) plots from gene set enrichment analysis (GSEA) using curated naive or primed pluripotency gene sets (see experimental procedures). Negative and positive normalized enrichment scores (NES) point to the gene set being over-represented in the top-most down- or upregulated genes in Stag1 KD mESCs, respectively. Vertical bars refer to individual genes in the gene set and their position reflects the contribution of each gene to the NES.
(G) Area occupied by APhi colonies relative to total colony area in mESCs treated with si scr and si SA1 from 3 independent replicates where n > 50 colonies/condition were counted.
(H) CRISPR-Cas9 was used to knock in a NeonGreen-v5-FKBP tag on both alleles of endogenous Stag1 at the C-terminus (SA1NG−FKBP). The resultant STAG1 protein is 42 kDa larger. Shown also are known features of STAG1 including the N-terminal AT-hook (AT) and the stromalin-conserved domain (SCD). WB analysis of STAG1 and NANOG levels in a targeted mESC clone after treatment with DMSO or dTAG. Tubulin (Tub) serves as a loading control.
(I) Area occupied by APhi colonies as above but in wild-type or SA1NG−FKBP mESCs treated with DMSO or dTAG. Data are from three independent replicates where n > 50 colonies/condition were counted. See also Figure S1.