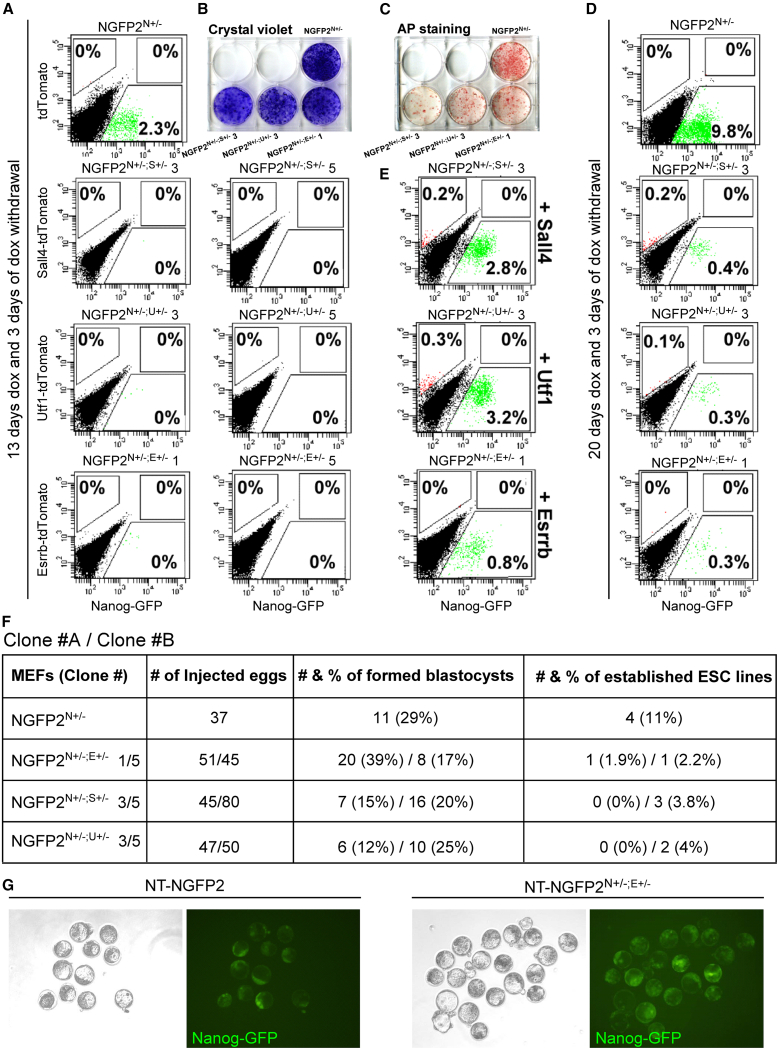
Figure 2.
NGFP2N+/− double heterozygous mutant MEFs show strong reprogramming inhibition either by OSKM or by NT
(A) Flow cytometry analysis of Nanog-GFP and tdTomato-positive cells for two different clones from each of the NGFP2N+/− double heterozygous mutant induced cells and control after 13 days of dox followed by 3 days of dox withdrawal. Representative flow cytometry plots are shown out of three independent reprogramming runs (n = 3).
(B and C) Crystal violet (B) and alkaline phosphatase (AP) (C) staining of whole reprogramming plates for each of the double heterozygous mutant induced line and control at the end of the reprogramming process. Representative stainings are depicted out of three independent reprogramming runs (n = 3).
(D) Flow cytometry analysis of Nanog-GFP and tdTomato-positive cells for each of the NGFP2N+/− double heterozygous mutant induced cells and control after 20 days of reprogramming. Representative flow cytometry plots are shown out of three independent reprogramming runs (n = 3).
(E) Flow cytometry analysis of Nanog-GFP and tdTomato-positive cells of each of the NGFP2N+/− double heterozygous mutant induced cells and control following overexpression of the targeted gene (Sall4, Utf1, and Esrrb) at the end of the reprogramming process. Representative flow cytometry plots are shown out of three independent reprogramming runs (n = 3).
(F) Table summarizes the efficiency (i.e., blastocyst formation and ESC derivation) of the NT experiments of MEF nuclei of the different double heterozygous mutant NGFP2N+/− lines. NGFP2N+/− (n = 37), NGFP2N+/−;E+/− (n = 96), NGFP2N+/−;s+/− (n = 125), and NGFP2N+/−;u+/− (n = 97). Numbers outside the “/” symbol indicate different targeted clones. For example, “1/5” represents clone #1 and clone #5 for the indicated system.
(G) Representative bright field and green channel images of NGFP2N+/− and NGFP2N+/−;E+/− after NT. See also Figure S2.