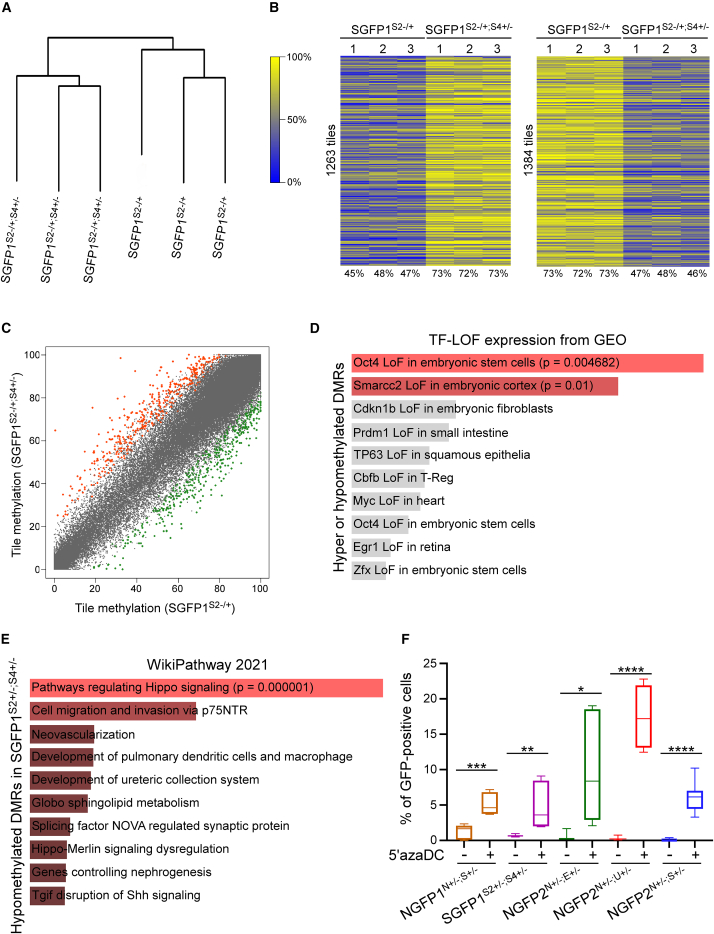
Figure 6.
DNA methylation abnormalities in the double heterozygous mutant fibroblasts hinder the reprogramming process
(A) Dendrogram for SGFP1S2+/− MEFs and SGFP1S2 +/-;S4+/− MEFs based on the level of relative change observed at CpG sites with a threshold of 10 reads per site. For each sample, three independent biological replicates are analyzed (n = 3).
(B) Heatmaps display DMRs (20%) in the indicated samples. Each tile (100 bp) is filtered to include at least 15 reads. p < 0.001. For each sample, three independent biological replicates are analyzed (n = 3).
(C) Scatterplot analysis (average of 3 replicates, n = 3) shows all the DMRs between SGFP1S2+/− MEFs and SGFP1S2+/-;S4+/− MEFs. Stained tiles are associated with genes that are related to pluripotency and development and are significantly more methylated in SGFP1S2+/-;S4+/− MEFs (red) or in SGFP1S2+/− MEFs (green).
(D and E) GO term enrichment analysis using different categories of EnrichR for hyper- or hypomethylated DMRs (D) or hypomethylated DMRs (E) in SGFP1S2+/-;S4+/− MEFs.
(F) Bar plot graph displays the percentage of GFP-positive cells in the indicated samples after 13 days of dox and 3 days of dox removal with and without prior treatment of 5′azaDC for two days. Boxes indicate 50% (25–75%) and whiskers (5–95%) of all measurements, with middle lines depicting the medians. Data are derived from six to nine independent reprogramming runs (n = 6–9). ∗p = 0.0127, ∗∗p = 0.0069, ∗∗∗p = 0.0001, ∗∗∗∗p < 0.0001 using a two-tailed unpaired t test calculated by GraphPad Prism (8.3.0). See also Figure S6.