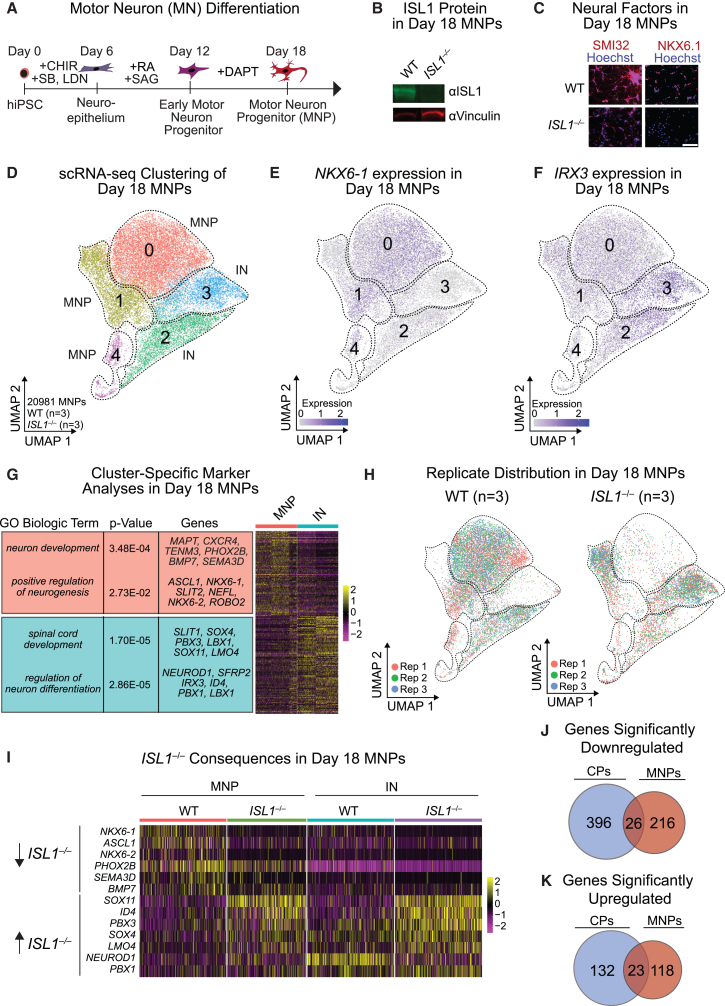
Figure 2.
scRNA-seq analyses of ISL1 function in MNPs
(A) Schematic of MN differentiation from hiPSCs.
(B) Western blot of ISL1 in WT and ISL1−/− day 18 MNPs. Vinculin served as the loading control.
(C) Immunofluorescence of neural factors SMI32 and NKX6.1 in day 18 MNPs. Scale bar, 100 μM.
(D) Hierarchical UMAP clustering of WT (n = 3 independent experiments) and ISL1−/− (n = 3 independent experiments) scRNA-seq data in day 18 iPSCs differentiated toward MNPs.
(E and F) Expression levels of the MN-related TF NKX6-1 (E) and the IN-related TF IRX3 (F) in day 18 MNPs, superimposed on the UMAP from (D).
(G) Heatmap of genes enriched with accompanying GO terms in each of the MNP or IN clusters depicted in (D).
(H) Replicate (Rep) comparison in WT (n = 3; 14,609 cells) or ISL1−/− (n = 3; 6,372 cells) day 18 MNPs across the clusters depicted in (D).
(I) Heatmap clustering of genes significantly dysregulated in scRNA-seq of ISL1−/−compared with WT day 18 MNPs.
(J) Venn diagram of significantly downregulated genes shared between ISL1−/− day 8 CPs and ISL1−/− day 18 MNPs.
(K) Venn diagram of significantly upregulated genes shared between ISL1−/− day 8 CPs and ISL1−/− day 18 MNPs.