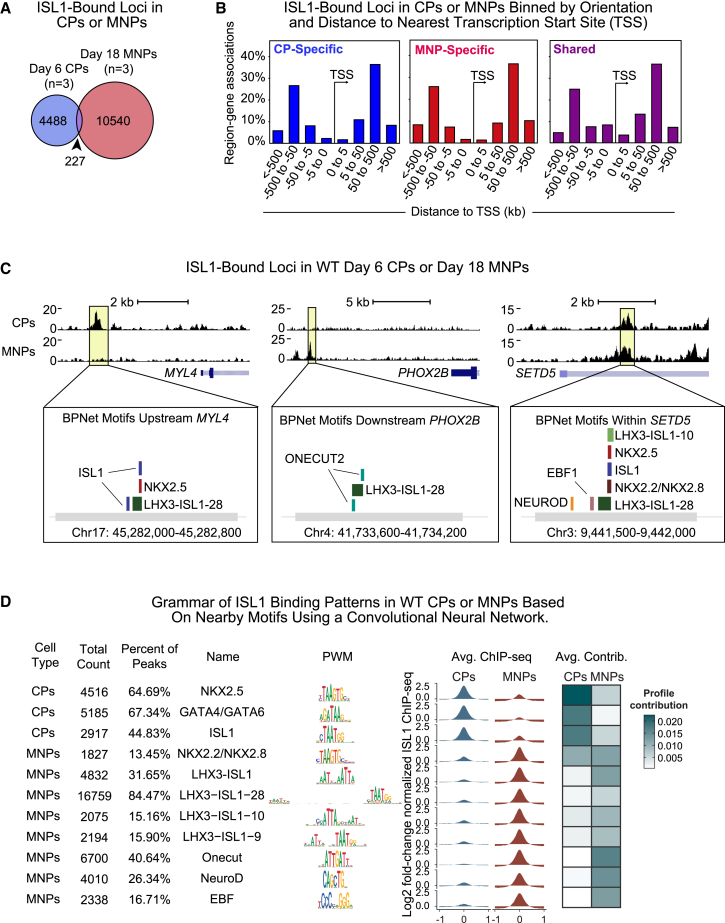
Figure 3.
Grammar of distinct ISL1 binding patterns in cardiac or MNPs based on nearby motifs using deep learning
(A) Venn diagram of ISL1 peaks in CPs or MNPs. Overlap is statistically significant (Regione R) (p = 0.001).
(B) Histogram of ISL1-bound peaks based on distance from gene TSSs.
(C) Example ChIP-seq tracks showing ISL1-bound loci with CP-specific, MNP-specific, or shared peaks. Corresponding de novo motifs identified by BPNet that predict cell-type-specific ISL1 binding are shown below. Data shown from a single representative replicate.
(D) De novo motifs identified by BPNet after simultaneous training of the CP-specific or MNP-specific binding profiles. Displayed for each identified motif is: the total count of motifs and percentages, name based on the most likely TF(s) that binds them, the position weight matrices (PWMs), and average ChIP-seq intensity and average contribution to the ISL1 binding predictions as extracted from BPNet.