Figure 4.
Proteomics and BPNet identify NKX2.5 and other TFs that guide ISL1 DNA-binding specificity in CPs
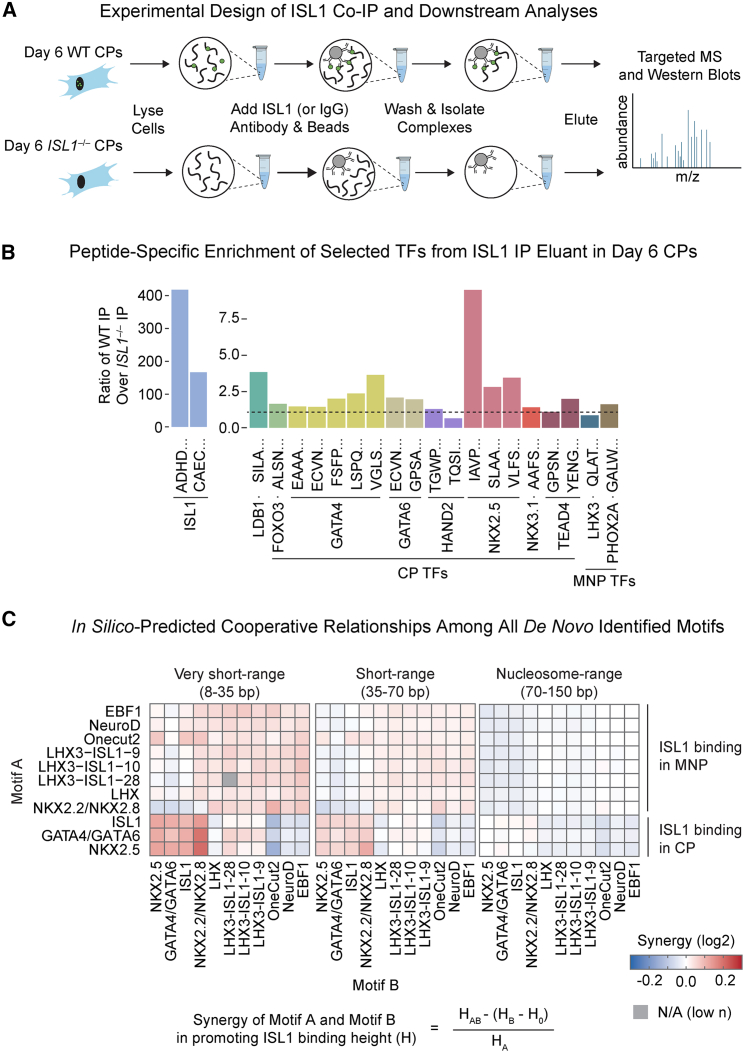
(A) Schematic for ISL1 co-IP and subsequent SRM targeted proteomics analyses.
(B) Barplot results from targeted affinity purification followed by MS, depicting enrichment of individual peptides corresponding to cardiac factors, positive controls (ISL1, LDB1), and negative controls (LHX3, PHOX2A) (n = 1 independent experiment).
(C) Cooperative relationship among all de novo identified motifs binned by distance from ISL1 binding predicted by in silico motif mutation.